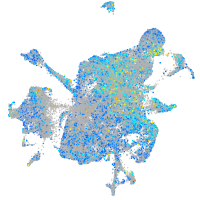
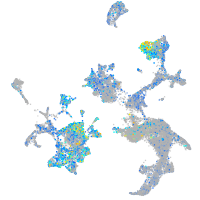
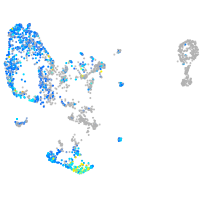
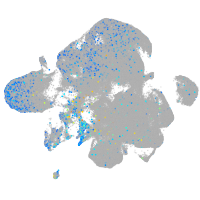
glutamate dehydrogenase 1a
ZFIN 
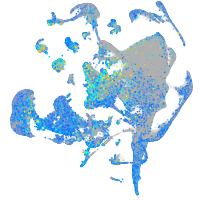
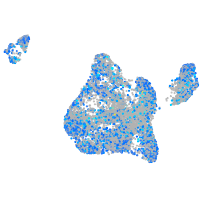
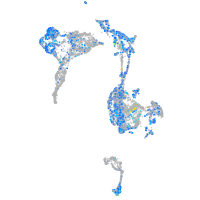
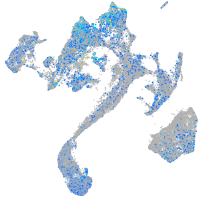
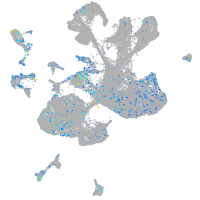
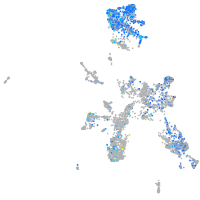
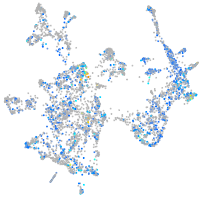
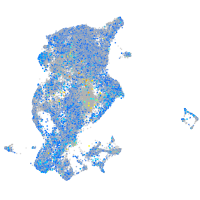
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:158423 | 0.488 | si:ch73-1a9.3 | -0.377 |
| cox6a2 | 0.477 | hspb1 | -0.372 |
| cox7a1 | 0.471 | hnrnpabb | -0.370 |
| slc7a8a | 0.469 | ptmab | -0.364 |
| si:ch211-139a5.9 | 0.458 | si:ch73-281n10.2 | -0.363 |
| sod2 | 0.455 | marcksb | -0.357 |
| aspg | 0.450 | h2afvb | -0.354 |
| atp1a1a.3 | 0.449 | wu:fb97g03 | -0.350 |
| trim35-12 | 0.447 | hmga1a | -0.349 |
| cdaa | 0.446 | si:ch211-222l21.1 | -0.348 |
| atp1b1a | 0.442 | nucks1a | -0.339 |
| vill | 0.441 | ilf3b | -0.328 |
| ccl19a.1 | 0.440 | hmgb2b | -0.326 |
| soul2 | 0.440 | hnrnpaba | -0.322 |
| chp1 | 0.438 | marcksl1b | -0.320 |
| ldhba | 0.437 | h3f3d | -0.310 |
| sfxn3 | 0.436 | snrpb | -0.309 |
| si:dkey-36i7.3 | 0.434 | seta | -0.307 |
| mal2 | 0.433 | cx43.4 | -0.305 |
| eef1da | 0.433 | apoeb | -0.304 |
| COX3 | 0.432 | hmgn6 | -0.304 |
| mt-atp6 | 0.432 | setb | -0.302 |
| mt-co2 | 0.432 | hmgn2 | -0.301 |
| ppp1r1b | 0.432 | syncrip | -0.300 |
| mt-nd2 | 0.430 | zgc:110425 | -0.299 |
| mt-nd4 | 0.426 | khdrbs1a | -0.298 |
| mt-nd1 | 0.425 | nono | -0.295 |
| slc25a5 | 0.425 | hmgb1b | -0.295 |
| ca4c | 0.424 | snrpg | -0.291 |
| cd63 | 0.423 | apoc1 | -0.288 |
| ponzr1 | 0.422 | ptges3b | -0.288 |
| mt-cyb | 0.417 | eif5a2 | -0.286 |
| tmem72 | 0.416 | lmo2 | -0.285 |
| cdh17 | 0.415 | snrpd2 | -0.285 |
| bin2a | 0.414 | ilf2 | -0.281 |