glycolipid transfer protein a
ZFIN 
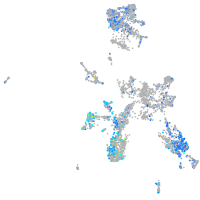
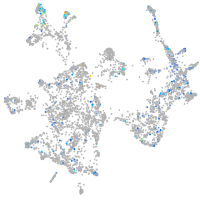
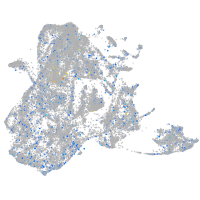
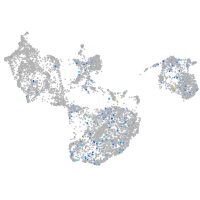
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cx30.3 | 0.215 | tyrp1b | -0.081 |
| sprb | 0.206 | tyrp1a | -0.075 |
| rgs2 | 0.190 | dct | -0.074 |
| pax7a | 0.182 | mdkb | -0.071 |
| mocos | 0.175 | ptmaa | -0.071 |
| mocs1 | 0.168 | anxa2a | -0.070 |
| cax1 | 0.165 | si:ch211-243a20.3 | -0.066 |
| si:dkeyp-110e4.6 | 0.162 | LOC103909099 | -0.066 |
| sigmar1 | 0.161 | oca2 | -0.066 |
| ppat | 0.159 | tuba1c | -0.066 |
| XLOC-016325 | 0.157 | zgc:91968 | -0.065 |
| pts | 0.156 | hmgb1b | -0.064 |
| slc2a11b | 0.155 | icn | -0.063 |
| prtfdc1 | 0.153 | s100a10b | -0.063 |
| pax7b | 0.148 | pmela | -0.063 |
| aox5 | 0.147 | prkar1b | -0.061 |
| XLOC-003678 | 0.141 | defbl1 | -0.059 |
| prdx1 | 0.140 | mtbl | -0.059 |
| impdh1b | 0.139 | hbae3 | -0.059 |
| mibp | 0.139 | nova2 | -0.059 |
| slc2a15a | 0.137 | apoda.1 | -0.058 |
| kctd12.2 | 0.135 | stmn1b | -0.057 |
| akr1b1 | 0.135 | slc24a5 | -0.057 |
| cyb5a | 0.132 | actc1b | -0.055 |
| gch2 | 0.131 | gng3 | -0.055 |
| CABZ01021592.1 | 0.129 | fxyd1 | -0.054 |
| gpd1b | 0.128 | tuba1a | -0.054 |
| glulb | 0.126 | hbbe1.3 | -0.054 |
| paics | 0.126 | pvalb1 | -0.053 |
| tmem130 | 0.124 | kita | -0.052 |
| bscl2l | 0.122 | spock3 | -0.052 |
| id3 | 0.121 | gpnmb | -0.052 |
| si:ch211-194k22.8 | 0.121 | fabp7a | -0.052 |
| dgkza | 0.121 | elavl3 | -0.052 |
| slc25a37 | 0.121 | gpm6aa | -0.051 |