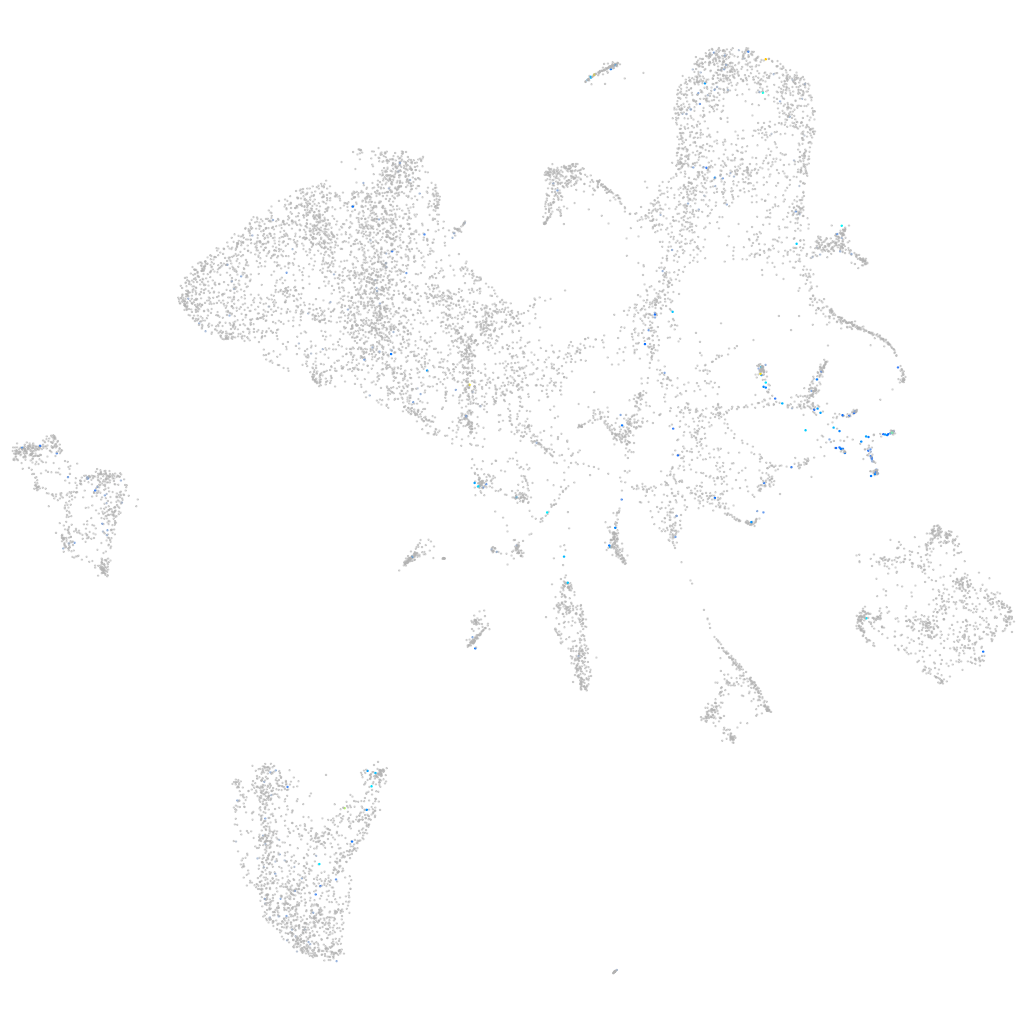
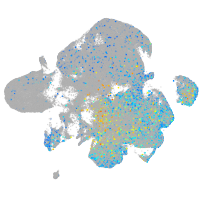
glutaminase a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.204 | aldob | -0.066 |
| trpm3 | 0.195 | ahcy | -0.065 |
| pax6b | 0.181 | aldh7a1 | -0.058 |
| si:dkey-153k10.9 | 0.180 | gamt | -0.058 |
| neurod1 | 0.178 | cebpd | -0.056 |
| kcnk9 | 0.175 | gapdh | -0.056 |
| gpr4 | 0.174 | apoc1 | -0.056 |
| pcsk1nl | 0.172 | si:dkey-16p21.8 | -0.056 |
| gnao1a | 0.172 | tfa | -0.056 |
| AP3B2 | 0.170 | serpina1l | -0.055 |
| scg2a | 0.168 | apoa1b | -0.055 |
| mir7a-1 | 0.164 | gstt1a | -0.055 |
| CABZ01095001.1 | 0.159 | fbp1b | -0.055 |
| vamp2 | 0.158 | shmt1 | -0.054 |
| pygma | 0.158 | mgst1.2 | -0.054 |
| map1b | 0.156 | cx32.3 | -0.054 |
| lysmd2 | 0.155 | cyp4v8 | -0.053 |
| slc35g2b | 0.155 | LOC110437731 | -0.053 |
| isl1 | 0.154 | eno3 | -0.053 |
| syt1a | 0.154 | apom | -0.053 |
| camk1da | 0.153 | serpina1 | -0.053 |
| NRXN2 | 0.153 | agxta | -0.053 |
| cacna1c | 0.152 | ces2 | -0.052 |
| aldocb | 0.151 | ttr | -0.052 |
| rnasekb | 0.150 | serpinf2b | -0.052 |
| insm1a | 0.149 | gc | -0.052 |
| clip3 | 0.146 | wu:fj16a03 | -0.052 |
| kcnj11 | 0.145 | fgg | -0.051 |
| UNC13A | 0.144 | fetub | -0.051 |
| atp6v0cb | 0.144 | nupr1b | -0.051 |
| tspan7b | 0.144 | apoa2 | -0.051 |
| myt1b | 0.144 | si:ch73-361h17.1 | -0.051 |
| pcsk1 | 0.143 | rgn | -0.051 |
| sypa | 0.142 | cfhl4 | -0.050 |
| scg2b | 0.141 | fgb | -0.050 |