"glutaminase 2b (liver, mitochondrial)"
ZFIN 
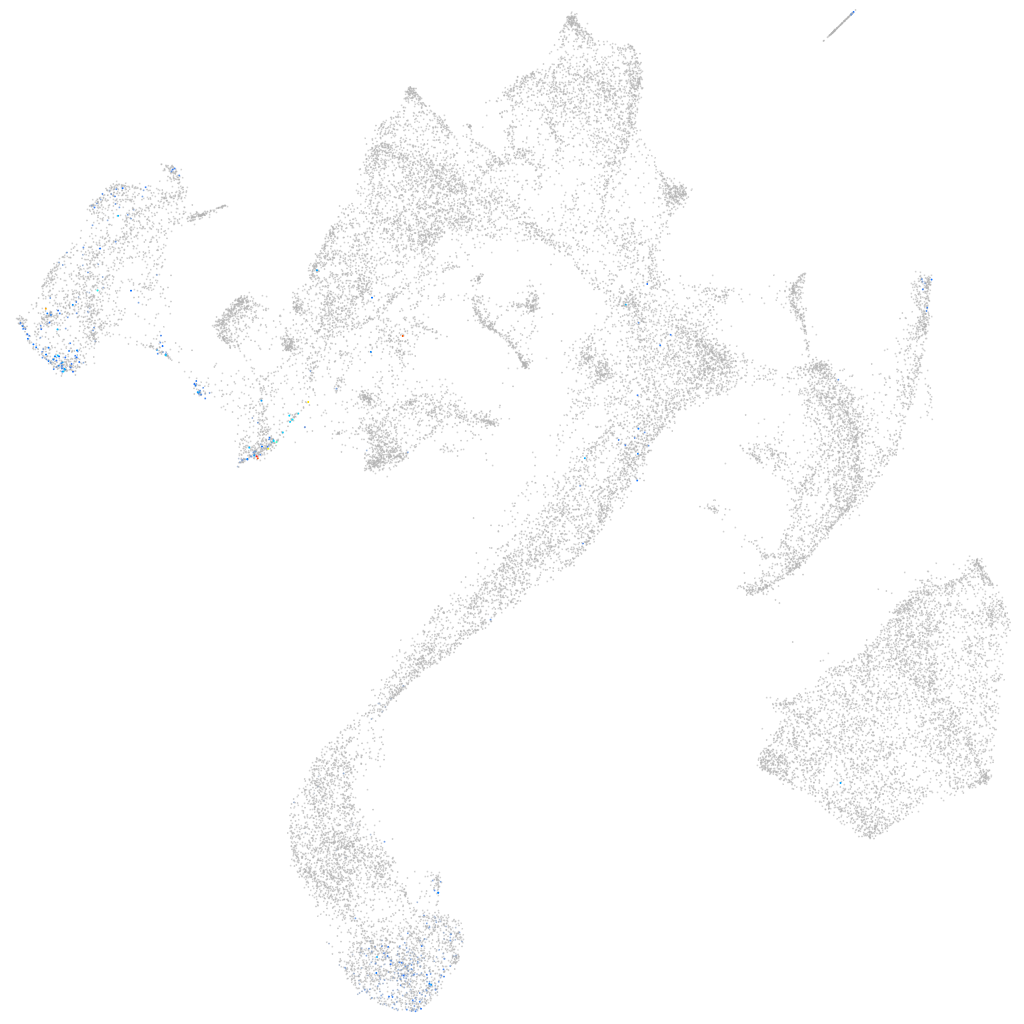
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU466278.2 | 0.243 | h3f3d | -0.144 |
| XLOC-041698 | 0.211 | hsp90ab1 | -0.134 |
| kcnt2 | 0.193 | rps12 | -0.132 |
| tnni2a.3 | 0.186 | naca | -0.128 |
| pvalb7 | 0.183 | cirbpb | -0.128 |
| ckmt2b | 0.177 | h2afvb | -0.127 |
| foxp3a | 0.177 | hnrnpaba | -0.127 |
| klhl38b | 0.171 | si:ch73-1a9.3 | -0.126 |
| atp2a2a | 0.171 | rplp2l | -0.126 |
| lpin1 | 0.168 | eef1a1l1 | -0.125 |
| pvalb4 | 0.167 | khdrbs1a | -0.124 |
| fam13b | 0.164 | rplp1 | -0.124 |
| wu:fj49a02 | 0.164 | pabpc1a | -0.121 |
| prx | 0.163 | rps16 | -0.120 |
| ache | 0.163 | ran | -0.119 |
| camk2a | 0.161 | rpl32 | -0.117 |
| si:ch211-266g18.10 | 0.161 | cirbpa | -0.117 |
| ankrd9 | 0.161 | hmga1a | -0.116 |
| si:ch211-195b15.7 | 0.161 | rpl7a | -0.114 |
| myom1b | 0.160 | si:ch211-222l21.1 | -0.113 |
| cox7a1 | 0.159 | snrpf | -0.112 |
| scn4ba | 0.158 | hmgn6 | -0.112 |
| ppp2r3a | 0.157 | hnrnpabb | -0.112 |
| CT027674.1 | 0.157 | rpl35 | -0.111 |
| nme2b.2 | 0.155 | eef2b | -0.111 |
| ablim2 | 0.155 | setb | -0.111 |
| got2a | 0.155 | hmgb2b | -0.111 |
| mylk4a | 0.153 | snrpb | -0.110 |
| CU681836.1 | 0.151 | ptmab | -0.109 |
| nrap | 0.150 | rpl36 | -0.108 |
| kbtbd12 | 0.149 | rpl13a | -0.107 |
| FO704758.2 | 0.149 | hnrnpa0b | -0.107 |
| ryr1a | 0.149 | rpl7 | -0.107 |
| obsl1b | 0.148 | cfl1 | -0.107 |
| si:dkeyp-50d11.2 | 0.148 | hmgn2 | -0.106 |