glyoxalase domain containing 5
ZFIN 
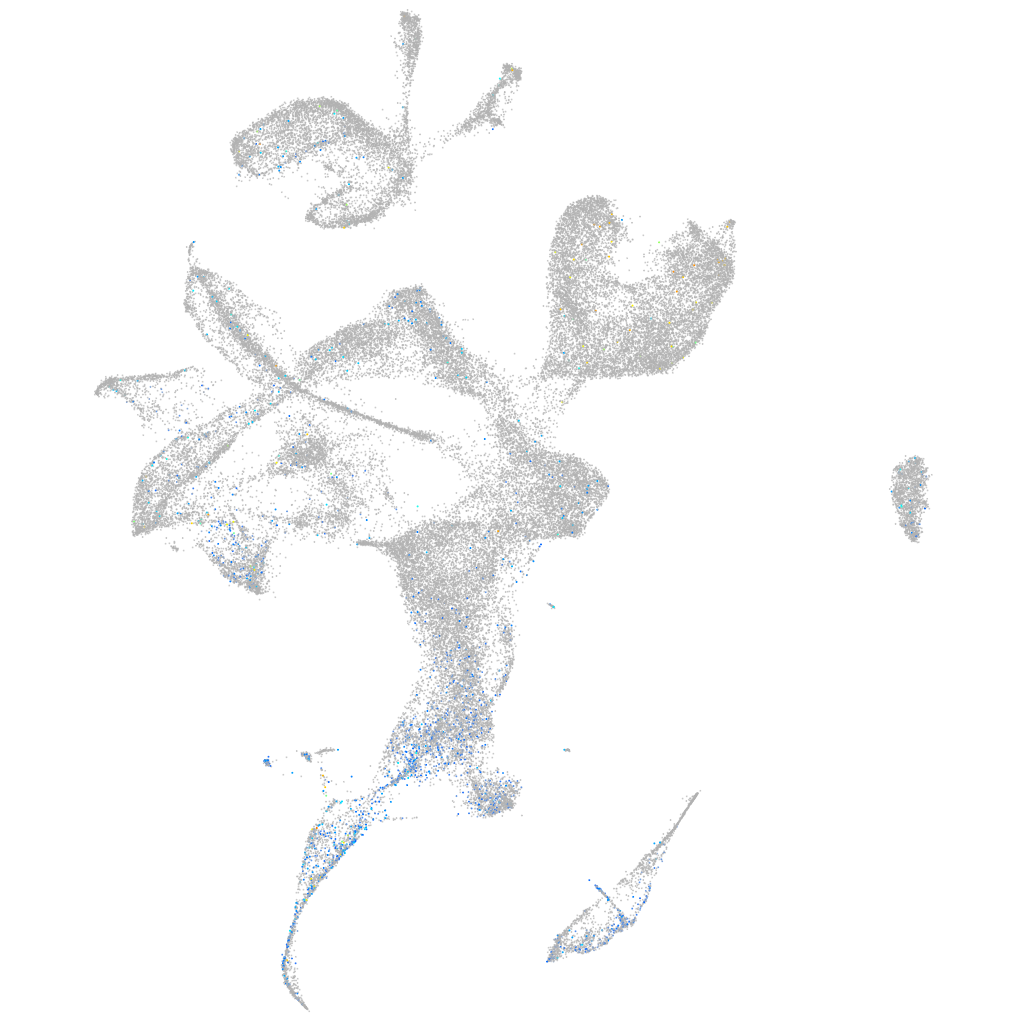
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tspan36 | 0.166 | ptmab | -0.115 |
| fabp11a | 0.165 | gpm6aa | -0.086 |
| pcbd1 | 0.156 | ptmaa | -0.082 |
| slc45a2 | 0.152 | si:ch73-1a9.3 | -0.077 |
| cx43 | 0.144 | hmgb1a | -0.076 |
| gpr143 | 0.142 | hmgn6 | -0.072 |
| dct | 0.142 | tuba1c | -0.070 |
| rab32a | 0.141 | gpm6ab | -0.070 |
| qdpra | 0.140 | marcksl1b | -0.065 |
| pmela | 0.139 | ckbb | -0.065 |
| zgc:110591 | 0.136 | tuba1a | -0.062 |
| agtrap | 0.134 | cadm3 | -0.060 |
| tyrp1b | 0.133 | nova2 | -0.058 |
| gstp1 | 0.132 | rorb | -0.057 |
| tyrp1a | 0.130 | btg1 | -0.056 |
| rab38 | 0.129 | epb41a | -0.053 |
| tfec | 0.128 | rtn1a | -0.052 |
| zgc:110239 | 0.128 | si:ch211-137a8.4 | -0.052 |
| sparc | 0.127 | neurod4 | -0.052 |
| FP085398.1 | 0.126 | stmn1b | -0.052 |
| rnaseka | 0.126 | foxg1b | -0.052 |
| trpm1b | 0.125 | scrt2 | -0.052 |
| adi1 | 0.124 | cspg5a | -0.051 |
| tmem88b | 0.123 | mdkb | -0.049 |
| tyr | 0.121 | tmem35 | -0.049 |
| mitfa | 0.117 | dbn1 | -0.048 |
| cd63 | 0.117 | hnrnpa0a | -0.047 |
| tspan10 | 0.116 | marcksl1a | -0.046 |
| wls | 0.115 | CR383676.1 | -0.046 |
| pmelb | 0.115 | chd4a | -0.046 |
| eif4ebp3l | 0.114 | fabp7a | -0.045 |
| sytl2b | 0.114 | myt1a | -0.045 |
| bace2 | 0.113 | vsx1 | -0.044 |
| sdc4 | 0.110 | anp32e | -0.044 |
| col4a5 | 0.109 | tp53inp2 | -0.044 |