golgi glycoprotein 1a
ZFIN 
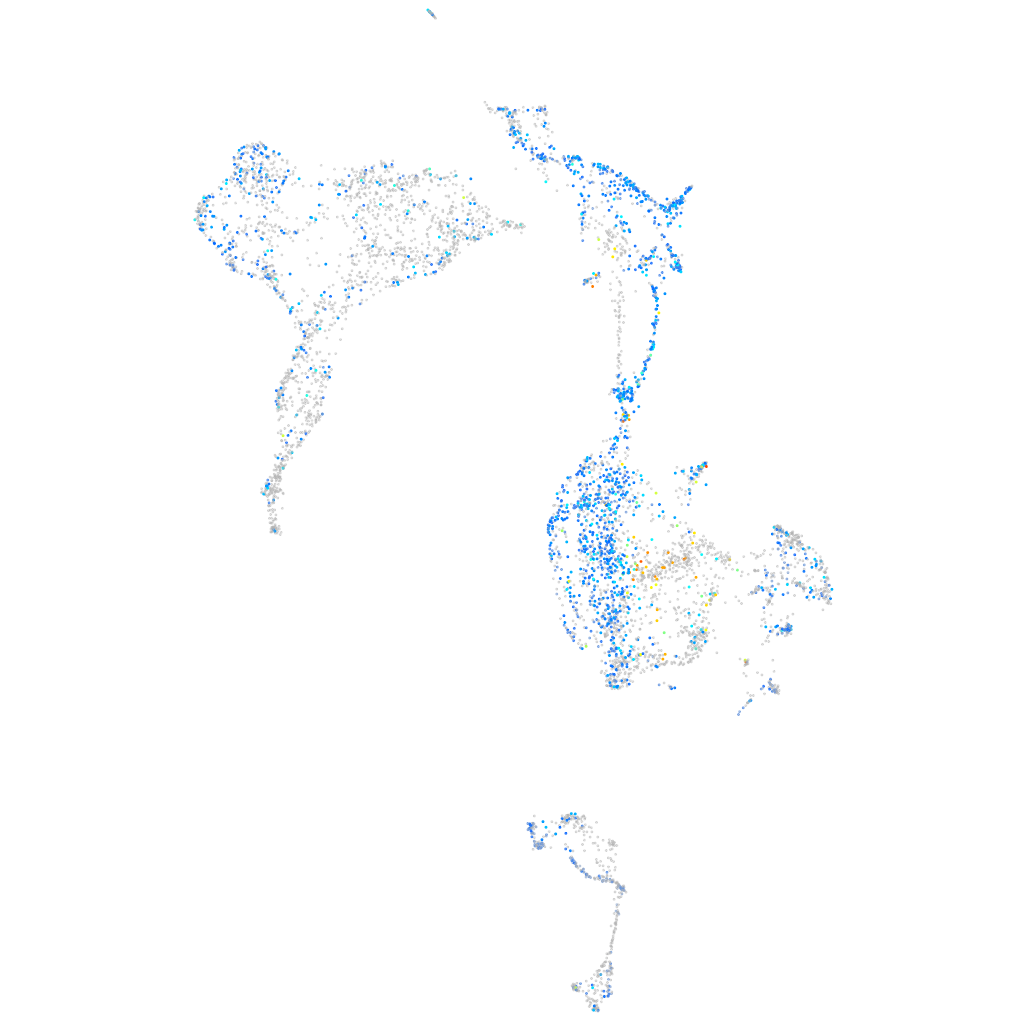
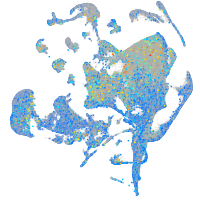
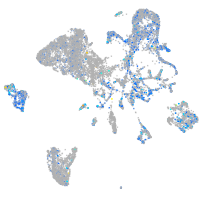
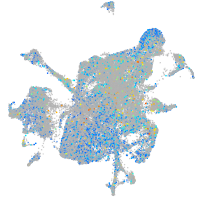
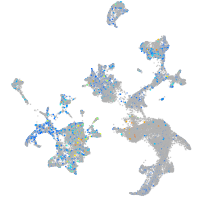
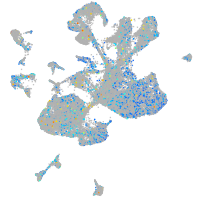
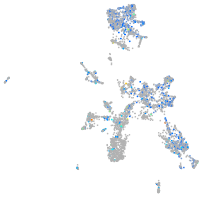
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| col2a1b | 0.328 | npm1a | -0.174 |
| col8a1a | 0.319 | nop56 | -0.168 |
| fkbp9 | 0.311 | nop58 | -0.168 |
| fkbp14 | 0.308 | fbl | -0.164 |
| plod3 | 0.295 | ncl | -0.163 |
| COLGALT1 (1 of many) | 0.292 | rsl1d1 | -0.160 |
| loxl3b | 0.292 | stm | -0.160 |
| ckap4 | 0.291 | hnrnpa1a | -0.155 |
| loxl1 | 0.289 | dkc1 | -0.154 |
| fkbp10b | 0.288 | ddx18 | -0.154 |
| kdelr3 | 0.279 | zmp:0000000624 | -0.154 |
| calua | 0.278 | nop2 | -0.154 |
| plod1a | 0.278 | hnrnpub | -0.153 |
| kdelr2b | 0.274 | hnrnpa1b | -0.153 |
| vkorc1 | 0.273 | mybbp1a | -0.151 |
| txndc5 | 0.273 | gnl3 | -0.150 |
| tgfb3 | 0.270 | ilf3b | -0.149 |
| p3h1 | 0.266 | si:ch211-222l21.1 | -0.148 |
| rcn1 | 0.266 | tardbp | -0.144 |
| col11a1a | 0.265 | rps18 | -0.141 |
| crtap | 0.264 | anp32e | -0.141 |
| lman1 | 0.260 | seta | -0.141 |
| rcn3 | 0.260 | fam50a | -0.141 |
| fkbp7 | 0.258 | ebna1bp2 | -0.138 |
| col4a5 | 0.257 | hnrnpabb | -0.138 |
| cyr61 | 0.255 | bms1 | -0.138 |
| CR381618.1 | 0.255 | rbmx2 | -0.137 |
| col2a1a | 0.253 | gar1 | -0.136 |
| col9a3 | 0.253 | fthl27 | -0.135 |
| slit3 | 0.252 | pes | -0.135 |
| mydgf | 0.252 | hnrnpd | -0.134 |
| cd81a | 0.249 | hspb1 | -0.133 |
| loxl2b | 0.249 | rrp15 | -0.132 |
| fstl1b | 0.249 | NC-002333.4 | -0.132 |
| itga6a | 0.247 | si:ch211-217k17.7 | -0.132 |