gamma-glutamyltransferase 5a
ZFIN 
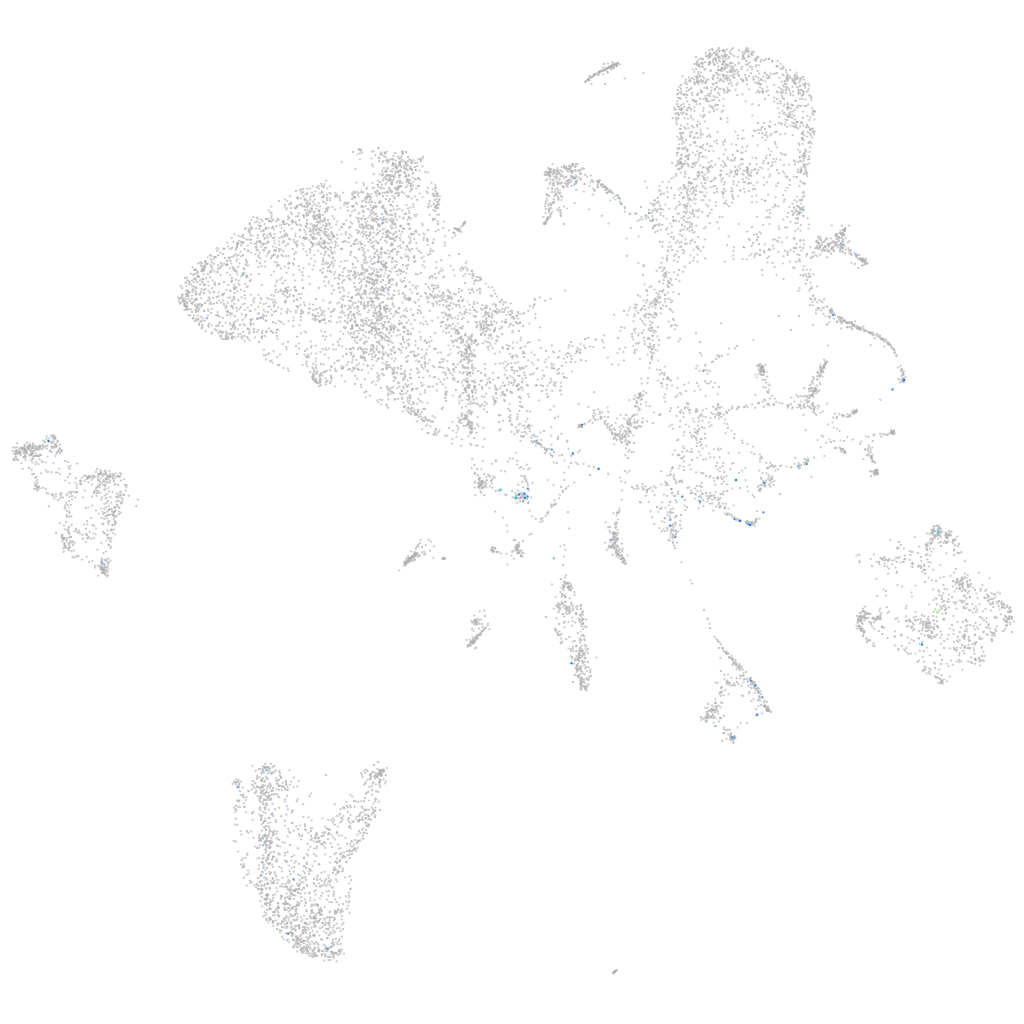
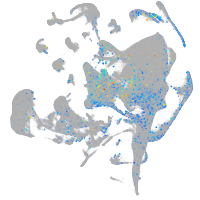
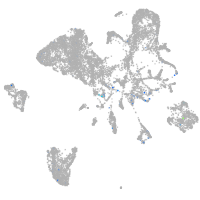
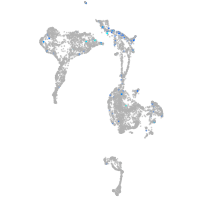
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX908388.2 | 0.330 | aldob | -0.064 |
| LOC100537584 | 0.308 | gapdh | -0.058 |
| XLOC-000357 | 0.248 | eno3 | -0.054 |
| zp2l2 | 0.217 | gpx4a | -0.052 |
| si:dkey-205i10.1 | 0.205 | tpi1b | -0.052 |
| aqp3b | 0.188 | fbp1b | -0.051 |
| tnfa | 0.186 | pklr | -0.050 |
| pax3b | 0.185 | apoc2 | -0.049 |
| XLOC-023858 | 0.183 | sod1 | -0.048 |
| CABZ01089980.1 | 0.169 | gstt1a | -0.048 |
| BX571719.1 | 0.158 | apoa4b.1 | -0.048 |
| pax3a | 0.157 | scp2a | -0.047 |
| LOC101885256 | 0.151 | gcshb | -0.047 |
| AL929536.5 | 0.150 | sult2st2 | -0.046 |
| FP245456.1 | 0.149 | ahcy | -0.045 |
| LOC108180305 | 0.148 | suclg1 | -0.045 |
| glt8d2 | 0.147 | gstr | -0.045 |
| LOC110439493 | 0.147 | mdh1aa | -0.045 |
| LO017718.1 | 0.141 | gamt | -0.044 |
| LOC103911713 | 0.140 | pgk1 | -0.044 |
| XLOC-003690 | 0.136 | mat1a | -0.044 |
| gli2a | 0.133 | nipsnap3a | -0.044 |
| CU657976.1 | 0.132 | apoa1b | -0.044 |
| LOC100000752 | 0.131 | glud1b | -0.044 |
| si:ch73-21g5.7 | 0.131 | ugt1a7 | -0.043 |
| cd37 | 0.128 | lgals2b | -0.043 |
| CT990561.1 | 0.127 | aldh6a1 | -0.043 |
| abhd6a | 0.127 | gpd1b | -0.042 |
| rfx4 | 0.127 | atp5mc3b | -0.042 |
| CABZ01078120.1 | 0.125 | ftcd | -0.042 |
| disc1 | 0.124 | pgam1a | -0.042 |
| LOC110439599 | 0.124 | grhprb | -0.042 |
| sulf2a | 0.122 | slco1d1 | -0.042 |
| XLOC-003689 | 0.121 | rdh1 | -0.041 |
| mir219-3 | 0.121 | si:dkey-16p21.8 | -0.041 |