"growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae)"
ZFIN 
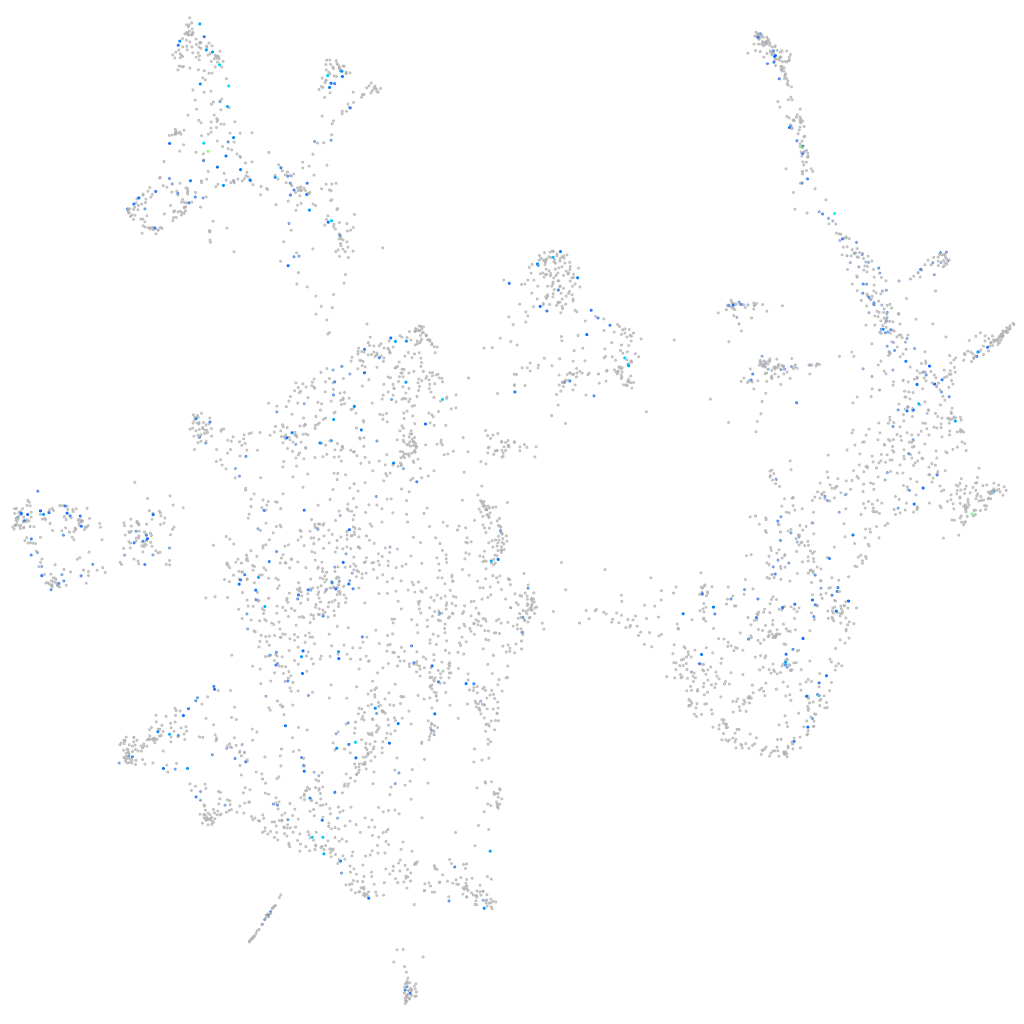
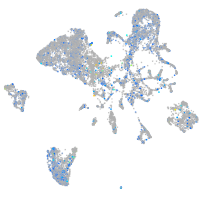
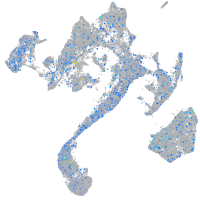
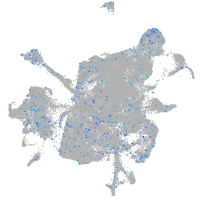
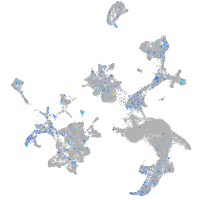
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dgat1a | 0.137 | CR383676.1 | -0.075 |
| angptl3 | 0.121 | rpl37 | -0.067 |
| aplnr2 | 0.119 | rpl38 | -0.067 |
| evpll | 0.115 | rpl29 | -0.063 |
| myo7aa | 0.114 | cd63 | -0.062 |
| calm3b | 0.113 | si:ch211-80h18.1 | -0.053 |
| LOC110437760 | 0.111 | sema3gb | -0.053 |
| ribc2 | 0.111 | pvalb2 | -0.053 |
| suclg1 | 0.111 | BX248331.2 | -0.050 |
| si:ch211-261p9.4 | 0.110 | tmsb4x | -0.050 |
| mfap3l | 0.108 | rps29 | -0.050 |
| gipc3 | 0.108 | rps21 | -0.049 |
| zgc:55461 | 0.108 | si:dkey-151g10.6 | -0.048 |
| cox6b1 | 0.106 | rps28 | -0.048 |
| tfg | 0.105 | otomp | -0.047 |
| cox6a1 | 0.105 | ppp1r1b | -0.046 |
| ap2m1a | 0.105 | sfrp5 | -0.046 |
| XLOC-044313 | 0.103 | ephb3 | -0.045 |
| arf2b | 0.102 | sparc | -0.045 |
| meig1 | 0.102 | rplp1 | -0.045 |
| LOC101885149 | 0.102 | slc12a2 | -0.044 |
| CABZ01055306.1 | 0.102 | tpma | -0.044 |
| myo6b | 0.102 | adh8b | -0.043 |
| uqcrfs1 | 0.101 | rpl39 | -0.042 |
| zgc:154006 | 0.100 | CABZ01086041.1 | -0.042 |
| dnah9l | 0.100 | prrg2 | -0.041 |
| tuba1a | 0.100 | col2a1b | -0.041 |
| parp6b | 0.100 | col15a1a | -0.041 |
| tmem107l | 0.100 | zgc:92313 | -0.041 |
| calm2a | 0.099 | dap1b | -0.040 |
| ccdc151 | 0.099 | si:ch211-39i22.1 | -0.040 |
| cox7a2a | 0.099 | gas1a | -0.040 |
| rab41 | 0.099 | col2a1a | -0.040 |
| catip | 0.099 | fbln7 | -0.040 |
| ap1s1 | 0.099 | ctrb1 | -0.039 |