gem (nuclear organelle) associated protein 6
ZFIN 
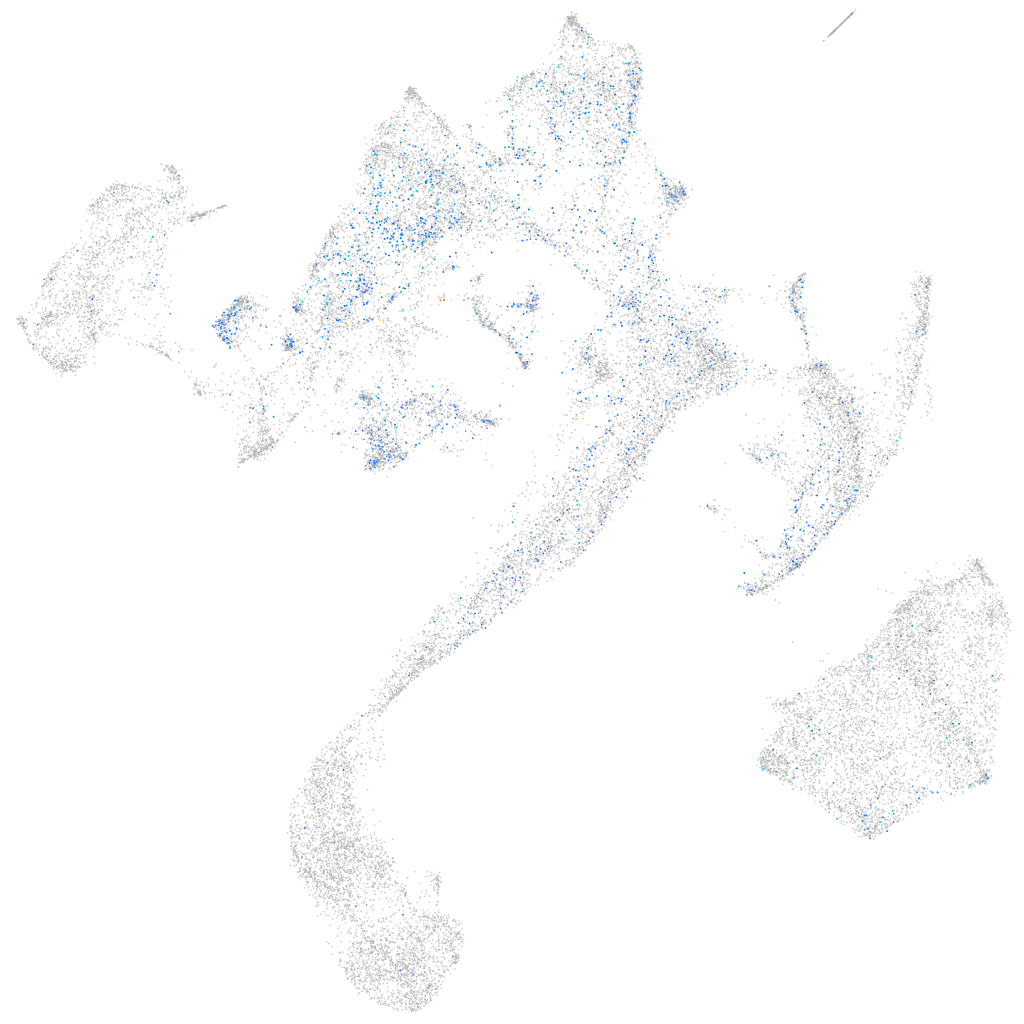
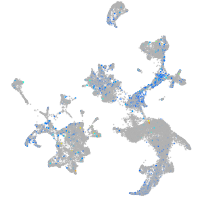
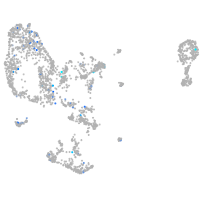
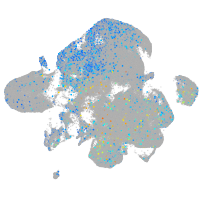
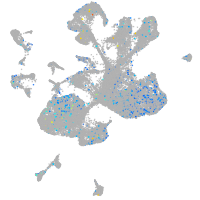
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-156b7.4 | 0.163 | ttn.1 | -0.102 |
| dut | 0.162 | tnnc2 | -0.098 |
| mif | 0.152 | hspb1 | -0.097 |
| h3f3a | 0.151 | neb | -0.097 |
| ppiab | 0.147 | gapdh | -0.096 |
| stmn1a | 0.147 | actn3b | -0.095 |
| zgc:56493 | 0.144 | aldoab | -0.095 |
| nme2b.1 | 0.143 | ak1 | -0.094 |
| prdx2 | 0.142 | smyd1a | -0.093 |
| pcna | 0.142 | ttn.2 | -0.092 |
| tuba8l3 | 0.134 | actn3a | -0.092 |
| hnrnpa0l | 0.132 | ldb3b | -0.092 |
| marcksl1a | 0.129 | myom1a | -0.092 |
| rnaseh2a | 0.129 | ckmb | -0.092 |
| ptmaa | 0.128 | atp2a1 | -0.091 |
| rrm2 | 0.127 | si:ch211-266g18.10 | -0.091 |
| prdx4 | 0.127 | mybphb | -0.091 |
| tuba8l | 0.126 | ldb3a | -0.091 |
| pmp22a | 0.125 | si:ch73-367p23.2 | -0.091 |
| nutf2l | 0.125 | tmod4 | -0.089 |
| tubb4b | 0.122 | pgam2 | -0.089 |
| eef1g | 0.121 | casq2 | -0.088 |
| eef1b2 | 0.121 | tpma | -0.088 |
| hspa8 | 0.121 | ckma | -0.088 |
| tuba8l4 | 0.120 | nme2b.2 | -0.087 |
| rps3a | 0.119 | hhatla | -0.087 |
| prim2 | 0.119 | XLOC-001975 | -0.087 |
| naa40 | 0.119 | prx | -0.087 |
| selenoh | 0.118 | ank1a | -0.086 |
| vwde | 0.118 | XLOC-025819 | -0.085 |
| rplp1 | 0.117 | acta1b | -0.085 |
| eif3ba | 0.117 | srl | -0.085 |
| rpa2 | 0.117 | CABZ01078594.1 | -0.084 |
| COX7A2 (1 of many) | 0.117 | casq1b | -0.084 |
| rpsa | 0.117 | tmem38a | -0.084 |