"glycine cleavage system protein H (aminomethyl carrier), a"
ZFIN 
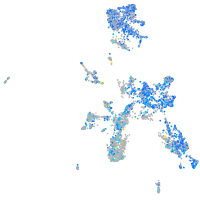
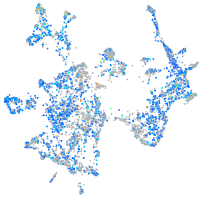
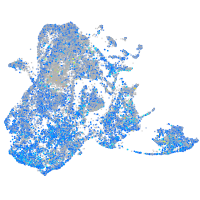
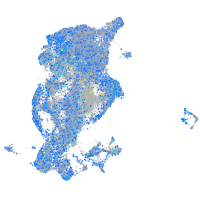
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cldn17 | 0.415 | si:dkey-67c22.2 | -0.328 |
| lzts2b | 0.389 | ccnt2b | -0.304 |
| tns2b | 0.387 | ing2 | -0.283 |
| srbd1 | 0.385 | nudc | -0.282 |
| lgals2a | 0.378 | prkrip1 | -0.276 |
| gipc2 | 0.377 | stk35l | -0.274 |
| unc5db | 0.372 | rhoab | -0.273 |
| tbl3 | 0.364 | tia1 | -0.269 |
| tcirg1a | 0.361 | eif4e1c | -0.268 |
| nfkbib | 0.361 | spink2.1 | -0.265 |
| ipo11 | 0.360 | nusap1 | -0.264 |
| rgcc | 0.358 | nkrf | -0.261 |
| khk | 0.355 | marcksl1b | -0.260 |
| LOC101884310 | 0.355 | eif2s1a | -0.258 |
| LO018148.1 | 0.355 | mfap1 | -0.258 |
| plekhg5a | 0.350 | si:dkeyp-69c1.6 | -0.257 |
| dnajc16 | 0.347 | si:dkey-208k4.2 | -0.250 |
| ttc7a | 0.347 | dctpp1 | -0.249 |
| nsmce2 | 0.346 | thoc7 | -0.248 |
| zgc:161969 | 0.346 | ncoa4 | -0.248 |
| gabarapa | 0.345 | lysmd3 | -0.247 |
| brms1la | 0.345 | si:dkey-27b3.2 | -0.243 |
| gmcl1 | 0.344 | pdcd4b | -0.241 |
| orai2 | 0.343 | tbx16 | -0.240 |
| cmtm4 | 0.343 | ammecr1 | -0.238 |
| zgc:92242 | 0.339 | rhebl1 | -0.235 |
| wscd2 | 0.338 | u2af2a | -0.235 |
| atxn7l2b | 0.335 | utp11l | -0.235 |
| tysnd1 | 0.335 | si:ch211-152c2.3 | -0.233 |
| aanat1 | 0.333 | ctsba | -0.233 |
| ephb6 | 0.328 | dhx37 | -0.232 |
| LOC101883330 | 0.325 | slc35f2 | -0.232 |
| si:ch211-246i5.5 | 0.325 | rrp12 | -0.231 |
| XLOC-010012 | 0.325 | ppfibp1b | -0.231 |
| tnfrsf19 | 0.325 | ddx18 | -0.229 |