germ cell nuclear acidic peptidase
ZFIN 
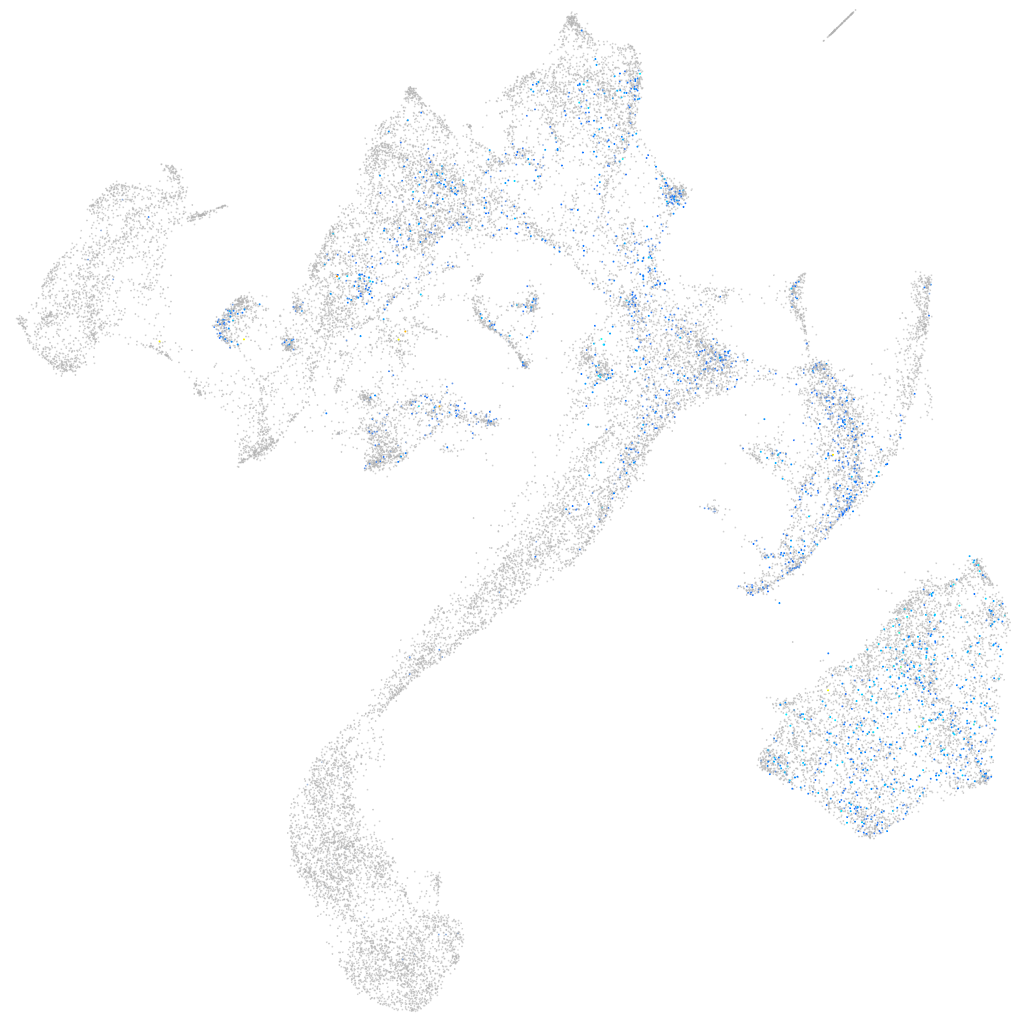
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdk1 | 0.300 | actc1b | -0.161 |
| tpx2 | 0.291 | aldoab | -0.144 |
| ube2c | 0.289 | ak1 | -0.140 |
| si:ch211-69g19.2 | 0.285 | tmem38a | -0.139 |
| plk1 | 0.282 | ckmb | -0.138 |
| cdc20 | 0.282 | ckma | -0.138 |
| kpna2 | 0.282 | atp2a1 | -0.137 |
| mki67 | 0.281 | ttn.2 | -0.135 |
| mad2l1 | 0.276 | ttn.1 | -0.135 |
| aurkb | 0.275 | tnnc2 | -0.134 |
| ccnb1 | 0.274 | acta1b | -0.133 |
| top2a | 0.272 | tpma | -0.131 |
| kif11 | 0.271 | desma | -0.130 |
| aspm | 0.271 | ldb3a | -0.129 |
| cdca8 | 0.269 | mybphb | -0.129 |
| tacc3 | 0.266 | acta1a | -0.128 |
| kifc1 | 0.257 | mylpfa | -0.128 |
| kif23 | 0.256 | CABZ01078594.1 | -0.128 |
| cenpf | 0.255 | neb | -0.128 |
| nusap1 | 0.255 | vdac3 | -0.127 |
| aurka | 0.252 | atp5if1b | -0.127 |
| kmt5ab | 0.250 | pabpc4 | -0.127 |
| ccna2 | 0.249 | eno1a | -0.126 |
| prc1a | 0.248 | srl | -0.126 |
| birc5a | 0.245 | cav3 | -0.124 |
| ccnb3 | 0.243 | ldb3b | -0.124 |
| dlgap5 | 0.243 | actn3b | -0.121 |
| g2e3 | 0.238 | gamt | -0.121 |
| kif20a | 0.233 | gapdh | -0.120 |
| spdl1 | 0.233 | nme2b.2 | -0.120 |
| armc1l | 0.229 | si:ch73-367p23.2 | -0.118 |
| cenpe | 0.228 | eno3 | -0.118 |
| hmgb2a | 0.227 | myl1 | -0.118 |
| numa1 | 0.224 | tnnt3a | -0.118 |
| kif22 | 0.223 | mylz3 | -0.117 |