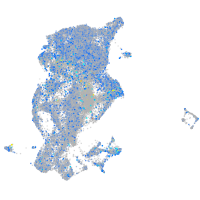
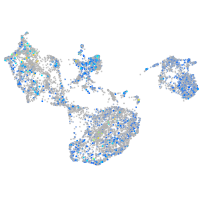
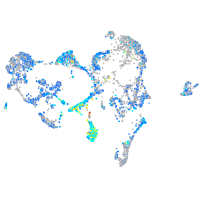
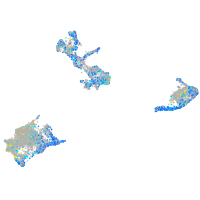
expressed sequence BC139872
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.315 | aqp12 | -0.147 |
| gapdhs | 0.272 | agxtb | -0.139 |
| pcsk1nl | 0.268 | bhmt | -0.132 |
| calm1b | 0.265 | gatm | -0.131 |
| neurod1 | 0.265 | gamt | -0.128 |
| fev | 0.246 | mat1a | -0.128 |
| atp6v1e1b | 0.245 | apoc1 | -0.126 |
| calm1a | 0.242 | pnp4b | -0.120 |
| si:ch1073-429i10.3.1 | 0.242 | gapdh | -0.120 |
| mir375-2 | 0.241 | fabp10a | -0.119 |
| vamp2 | 0.241 | ttr | -0.118 |
| rnasekb | 0.238 | uox | -0.116 |
| scgn | 0.237 | agxta | -0.115 |
| c2cd4a | 0.236 | ttc36 | -0.114 |
| hepacam2 | 0.234 | cpn1 | -0.114 |
| si:dkey-153k10.9 | 0.234 | ahcy | -0.114 |
| egr4 | 0.232 | rbp2b | -0.112 |
| syt1a | 0.230 | aldh6a1 | -0.111 |
| pcsk1 | 0.230 | gc | -0.110 |
| pax6b | 0.230 | ambp | -0.110 |
| mir7a-1 | 0.229 | ppdpfa | -0.110 |
| capns1b | 0.228 | nupr1b | -0.110 |
| ier2b | 0.228 | hpda | -0.109 |
| atp6ap2 | 0.226 | ftcd | -0.109 |
| id4 | 0.225 | ces2 | -0.109 |
| sat1a.2 | 0.224 | serpinf2b | -0.108 |
| tmem59 | 0.223 | apoa1b | -0.108 |
| rprmb | 0.221 | ssr1 | -0.108 |
| elovl1a | 0.218 | ugp2a | -0.107 |
| cd63 | 0.218 | LOC110437731 | -0.107 |
| zgc:101731 | 0.218 | si:dkey-86l18.10 | -0.107 |
| vat1 | 0.217 | serpina1l | -0.107 |
| insm1a | 0.217 | aldh7a1 | -0.106 |
| oaz1b | 0.216 | c9 | -0.105 |
| scg2a | 0.215 | zgc:123103 | -0.105 |