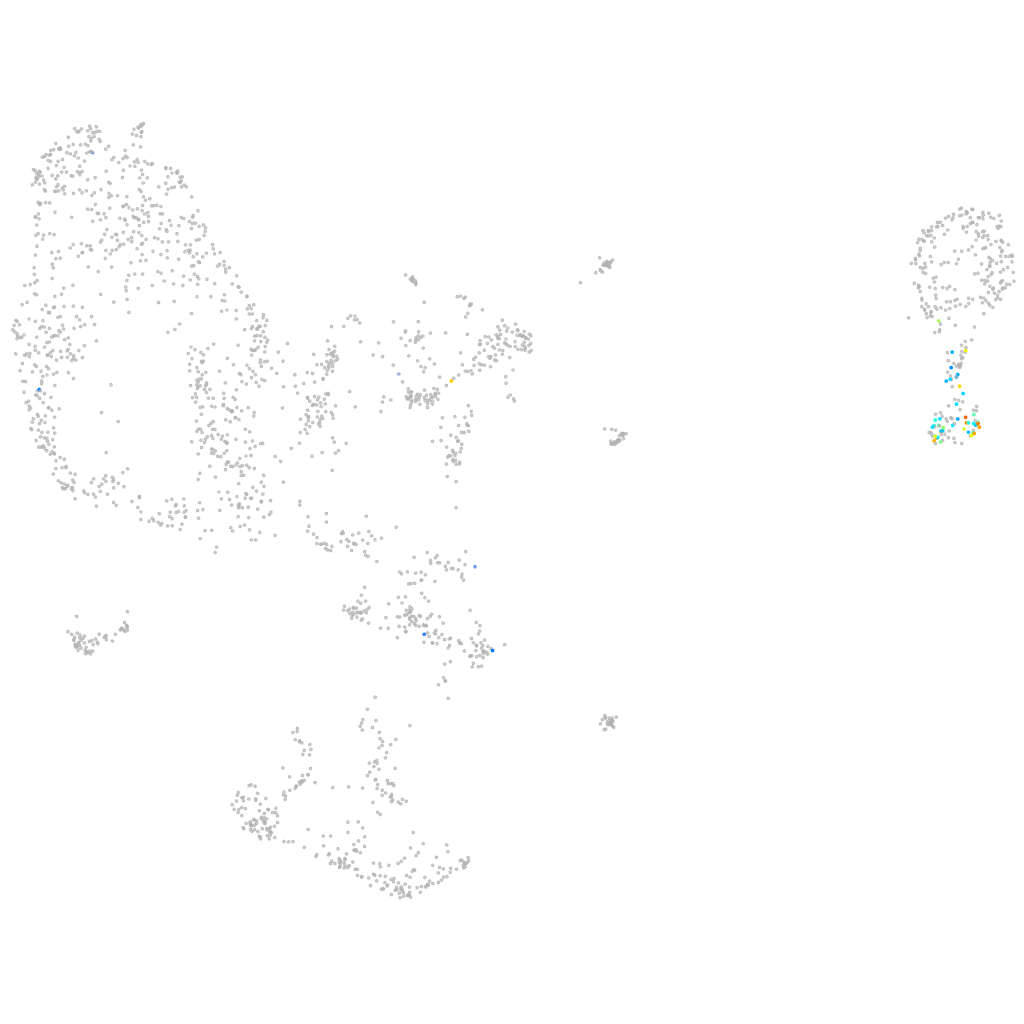
GATA binding protein 1a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 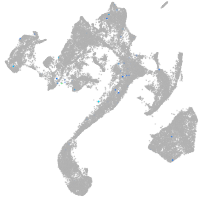 |
mural cells / non-skeletal muscle 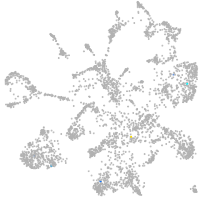 |
mesenchyme 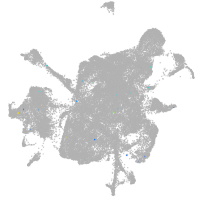 |
hematopoietic / vasculature 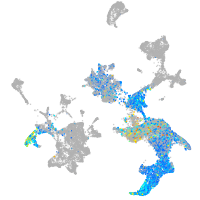 |
pronephros 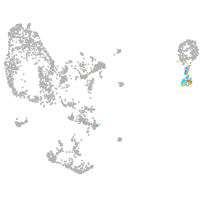 |
neural |
spinal cord / glia |
eye |
taste / olfactory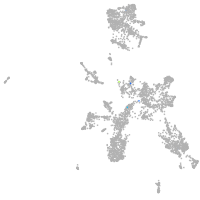 |
otic / lateral line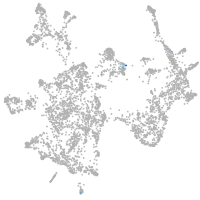 |
epidermis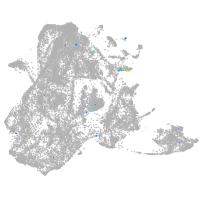 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gfi1aa | 0.673 | rpl37 | -0.304 |
| klf17 | 0.648 | rps10 | -0.303 |
| morc3b | 0.595 | zgc:114188 | -0.283 |
| si:ch73-299h12.2 | 0.539 | rps17 | -0.259 |
| tal1 | 0.469 | zgc:56493 | -0.232 |
| hhex | 0.463 | atp5f1b | -0.207 |
| lmo2 | 0.459 | atp5mc1 | -0.202 |
| cldng | 0.368 | ndufa4l | -0.200 |
| si:ch73-299h12.3 | 0.364 | cox4i1 | -0.199 |
| sox7 | 0.359 | mdh1aa | -0.198 |
| runx1 | 0.350 | slc25a5 | -0.197 |
| wu:fb97g03 | 0.327 | COX5B | -0.196 |
| mphosph10 | 0.317 | actb2 | -0.194 |
| lcp2b | 0.315 | suclg1 | -0.193 |
| mybbp1a | 0.308 | vdac3 | -0.193 |
| pprc1 | 0.308 | ndrg1a | -0.188 |
| ripply2 | 0.305 | ldhba | -0.188 |
| rbmx2 | 0.293 | cdh17 | -0.186 |
| flrt3 | 0.292 | idh2 | -0.186 |
| npas4l | 0.288 | atp1b1a | -0.184 |
| ncl | 0.287 | atp5mc3b | -0.183 |
| gnl3 | 0.287 | hnrnpa0l | -0.182 |
| lyar | 0.282 | atp5fa1 | -0.181 |
| pwp1 | 0.281 | h3f3a | -0.179 |
| DIPK2B | 0.281 | eno3 | -0.176 |
| nop2 | 0.279 | COX7A2 (1 of many) | -0.175 |
| nip7 | 0.278 | zgc:158463 | -0.173 |
| drl | 0.273 | atp5l | -0.172 |
| nufip1 | 0.273 | si:ch211-139a5.9 | -0.171 |
| npm1a | 0.273 | atp5if1b | -0.170 |
| dkc1 | 0.272 | dap1b | -0.169 |
| nop58 | 0.269 | si:dkey-16p21.8 | -0.169 |
| fbl | 0.267 | mt-atp6 | -0.168 |
| mak16 | 0.265 | tpi1b | -0.167 |
| pinx1 | 0.265 | ppdpfb | -0.164 |