"growth arrest and DNA-damage-inducible, gamma b, tandem duplicate 1"
ZFIN 
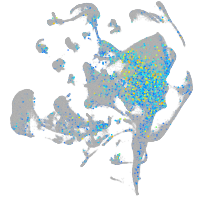
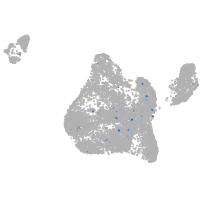
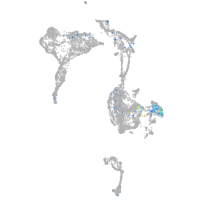
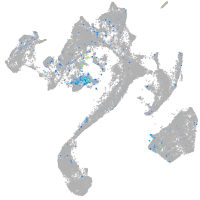
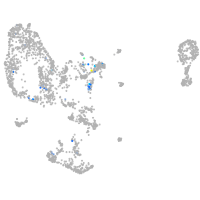
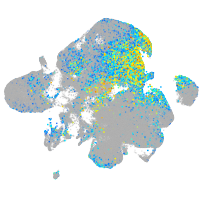
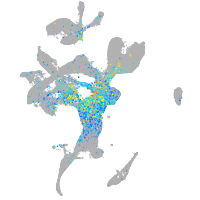
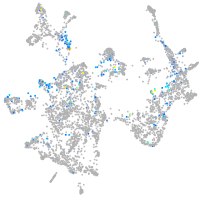
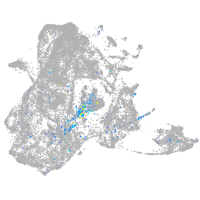
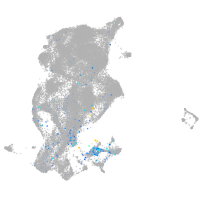
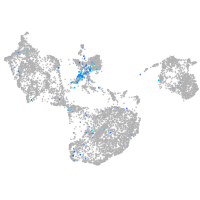
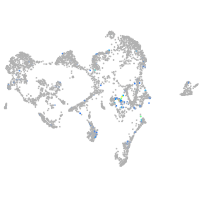
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-003692 | 0.374 | lmo2 | -0.059 |
| dla | 0.372 | mcl1a | -0.056 |
| dlb | 0.360 | pfn1 | -0.050 |
| abhd6a | 0.334 | rac2 | -0.042 |
| her4.2 | 0.334 | cebpb | -0.042 |
| XLOC-003689 | 0.317 | laptm5 | -0.042 |
| XLOC-003690 | 0.316 | wasb | -0.039 |
| insm1a | 0.310 | wasa | -0.039 |
| LOC798783 | 0.310 | coro1a | -0.038 |
| her4.1 | 0.297 | grap2b | -0.038 |
| CU634008.1 | 0.294 | si:dkey-262k9.4 | -0.038 |
| her4.4 | 0.291 | gmfg | -0.038 |
| BX530077.1 | 0.281 | clic1 | -0.037 |
| her15.1 | 0.280 | clic2 | -0.037 |
| CU467822.1 | 0.276 | rgs13 | -0.037 |
| her13 | 0.272 | si:ch73-248e21.7 | -0.037 |
| im:7152348 | 0.267 | arpc1b | -0.037 |
| elavl3 | 0.263 | glrx | -0.037 |
| si:ch73-21g5.7 | 0.256 | dusp5 | -0.037 |
| neurod4 | 0.249 | srgn | -0.037 |
| ebf2 | 0.241 | eno3 | -0.037 |
| sox11b | 0.240 | hhex | -0.036 |
| myt1b | 0.236 | crip1 | -0.036 |
| scrt2 | 0.228 | sat1a.2 | -0.036 |
| mdkb | 0.226 | hmbsa | -0.035 |
| myt1a | 0.225 | znfl2a | -0.035 |
| fabp7a | 0.223 | atp1b1a | -0.035 |
| hes6 | 0.222 | si:dkey-112e7.2 | -0.034 |
| si:ch211-193l2.6 | 0.208 | spi1b | -0.034 |
| si:ch211-137a8.4 | 0.208 | slc11a2 | -0.034 |
| tuba1a | 0.205 | litaf | -0.034 |
| neurog1 | 0.204 | ccr9a | -0.034 |
| msi1 | 0.203 | itm2bb | -0.034 |
| hmgb1b | 0.203 | si:dkey-261j4.4 | -0.034 |
| ascl1a | 0.200 | rnaseka | -0.034 |