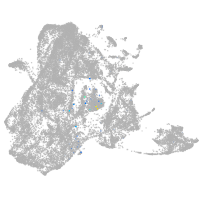
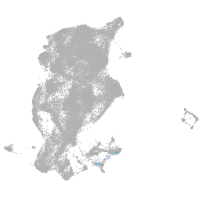
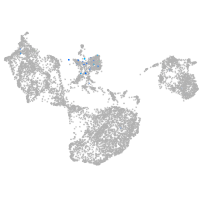
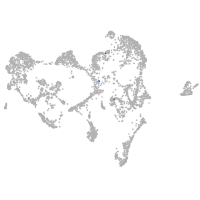
"gamma-aminobutyric acid (GABA) A receptor, zeta"
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| MGAT5B | 0.704 | rpl30 | -0.086 |
| col8a1b | 0.683 | rpl13 | -0.067 |
| LO018303.1 | 0.679 | serbp1a | -0.066 |
| XLOC-044313 | 0.626 | eif3ba | -0.064 |
| LOC108181592 | 0.560 | rpl11 | -0.060 |
| ephb1 | 0.557 | hspe1 | -0.056 |
| CEP170B (1 of many) | 0.536 | eif2s2 | -0.055 |
| LOC100334937 | 0.534 | eif3f | -0.054 |
| erbb4a | 0.523 | ndrg3a | -0.054 |
| slco3a1 | 0.507 | rpl18a | -0.053 |
| angpt2a | 0.506 | prkab1b | -0.053 |
| si:dkeyp-72e1.9 | 0.459 | rpl29 | -0.053 |
| si:ch211-241b2.4 | 0.453 | rps13 | -0.052 |
| cadm1b | 0.438 | cd9b | -0.051 |
| cd248a | 0.437 | anxa4 | -0.050 |
| myo10l1 | 0.436 | tuba8l | -0.050 |
| map3k9 | 0.424 | tuba8l2 | -0.049 |
| LOC103909205 | 0.413 | STMP1 | -0.049 |
| slc7a4 | 0.407 | rps6 | -0.049 |
| rab39ba | 0.403 | eif2s3 | -0.049 |
| gpr27 | 0.403 | zgc:86598 | -0.049 |
| clvs2 | 0.402 | rpl19 | -0.048 |
| inhbab | 0.401 | btf3 | -0.047 |
| XLOC-011941 | 0.398 | bzw1b | -0.047 |
| csf1b | 0.394 | fosab | -0.047 |
| LO017700.2 | 0.392 | zgc:110339 | -0.047 |
| LOC110437922 | 0.392 | csde1 | -0.047 |
| LOC797394 | 0.392 | chp1 | -0.046 |
| SLC29A4 (1 of many) | 0.389 | ppp1cbl | -0.046 |
| LOC108191314 | 0.384 | rps4x | -0.046 |
| esyt1a | 0.379 | eif3i | -0.046 |
| sgk494b | 0.372 | ak2 | -0.045 |
| slc25a51a | 0.369 | hdlbpa | -0.045 |
| BX001023.1 | 0.368 | selenoh | -0.044 |
| CABZ01088036.1 | 0.362 | rnaseka | -0.044 |