"fucosyltransferase 7 (alpha (1,3) fucosyltransferase)"
ZFIN 
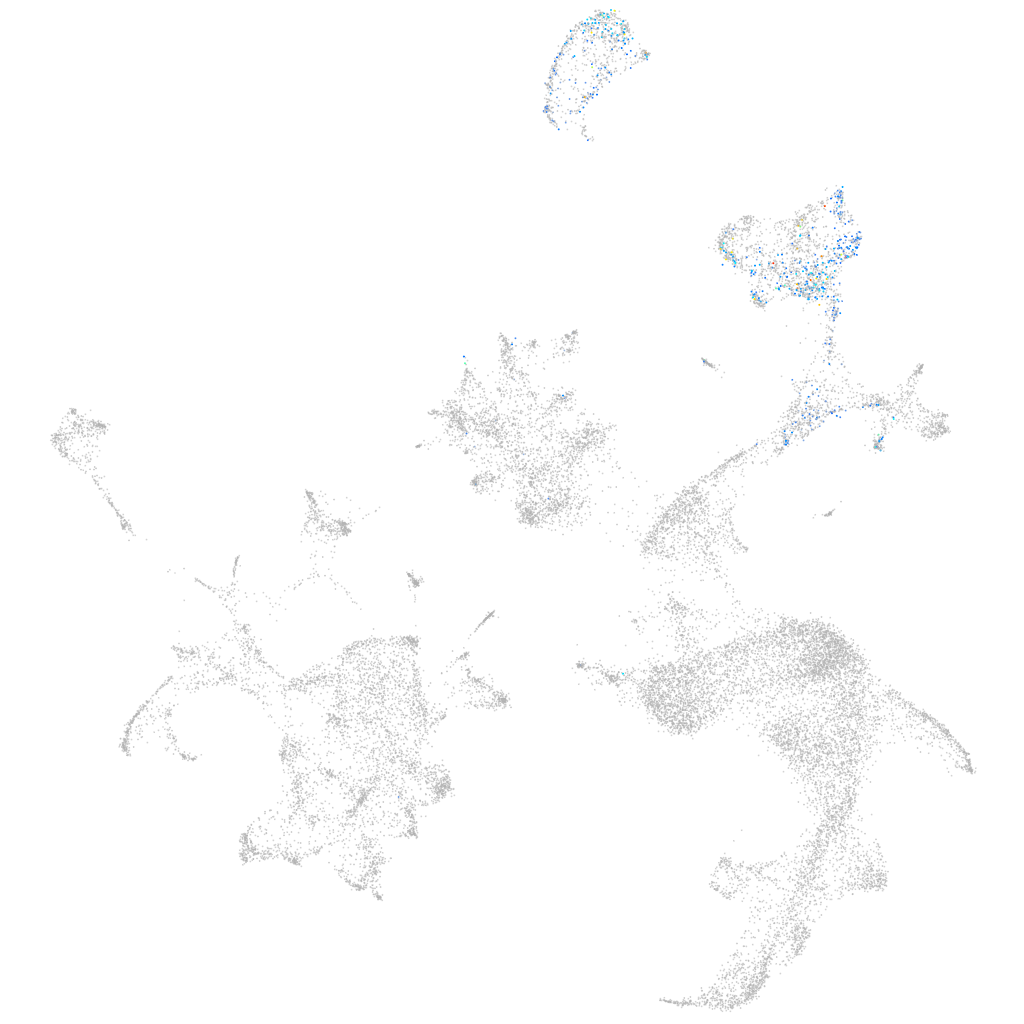
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fcer1gl | 0.298 | hbae1.1 | -0.125 |
| il1b | 0.280 | hbbe1.3 | -0.124 |
| ctss2.1 | 0.278 | hbae3 | -0.124 |
| spi1b | 0.271 | hbae1.3 | -0.122 |
| laptm5 | 0.268 | hbbe2 | -0.121 |
| coro1a | 0.268 | hbbe1.1 | -0.116 |
| cyba | 0.266 | cahz | -0.112 |
| plxnc1 | 0.263 | hbbe1.2 | -0.105 |
| arpc1b | 0.260 | hemgn | -0.104 |
| si:ch211-102c2.4 | 0.255 | fth1a | -0.100 |
| rac2 | 0.255 | alas2 | -0.098 |
| zgc:64051 | 0.254 | zgc:56095 | -0.097 |
| glipr1a | 0.251 | si:ch211-250g4.3 | -0.096 |
| cebpb | 0.251 | lmo2 | -0.093 |
| CABZ01078737.1 | 0.244 | creg1 | -0.093 |
| ptpn6 | 0.242 | slc4a1a | -0.092 |
| tnfb | 0.239 | nt5c2l1 | -0.089 |
| mfap4 | 0.238 | blvrb | -0.087 |
| capgb | 0.238 | epb41b | -0.086 |
| cmklr1 | 0.238 | zgc:163057 | -0.086 |
| vsir | 0.237 | si:ch211-207c6.2 | -0.081 |
| cotl1 | 0.233 | aqp1a.1 | -0.079 |
| havcr1 | 0.233 | nmt1b | -0.079 |
| ncf1 | 0.232 | tspo | -0.079 |
| itgb2 | 0.232 | plac8l1 | -0.076 |
| si:dkey-5n18.1 | 0.232 | si:ch211-227m13.1 | -0.070 |
| acod1 | 0.231 | tmod4 | -0.070 |
| si:dkey-102g19.3 | 0.231 | sptb | -0.070 |
| lygl1 | 0.231 | hbae5 | -0.069 |
| hcls1 | 0.229 | uros | -0.069 |
| nrros | 0.229 | znfl2a | -0.069 |
| cxcr3.2 | 0.228 | arl4aa | -0.068 |
| si:ch73-343l4.2 | 0.227 | histh1l | -0.068 |
| mapre1a | 0.227 | tfr1a | -0.067 |
| ctss2.2 | 0.226 | rfesd | -0.066 |