"fucosyltransferase 11 (alpha (1,3) fucosyltransferase)"
ZFIN 
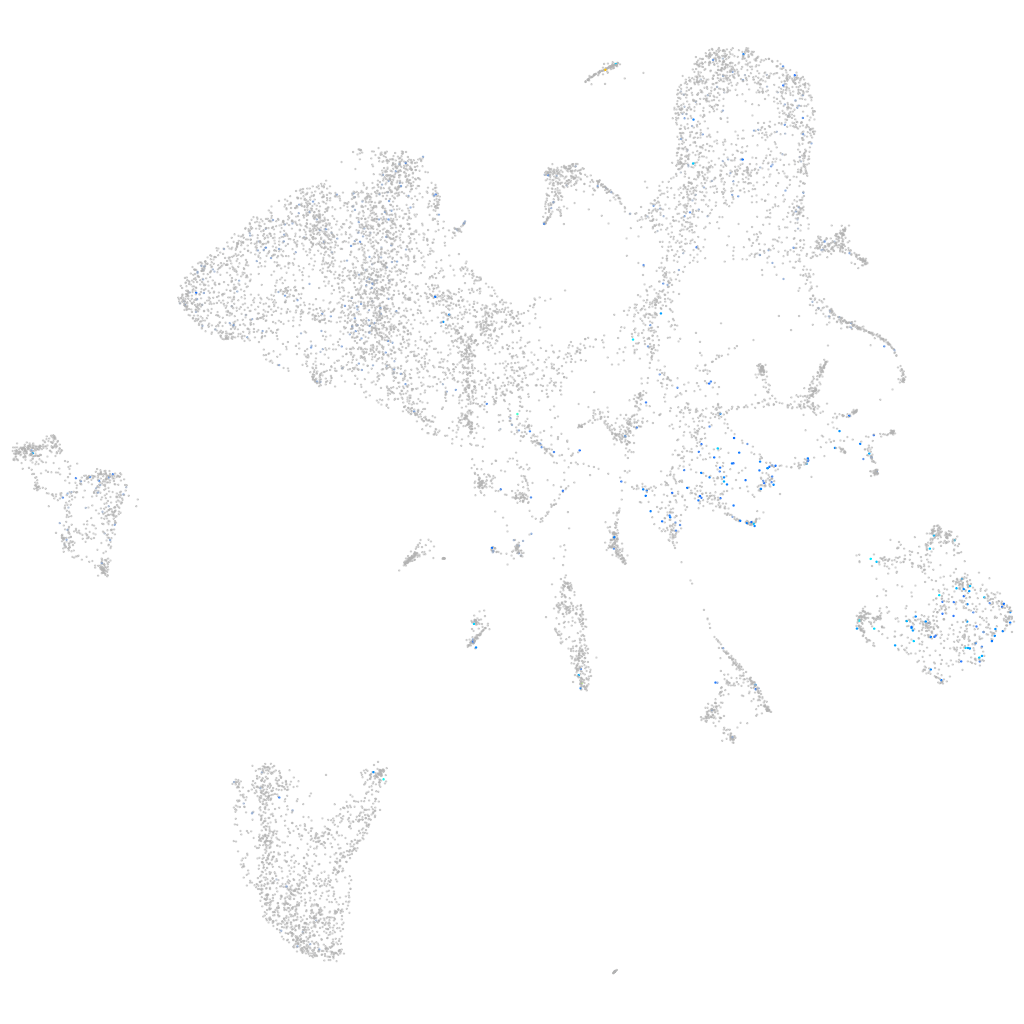
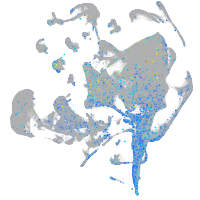
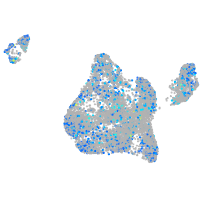
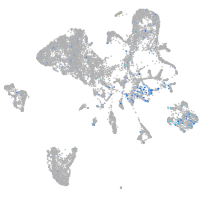
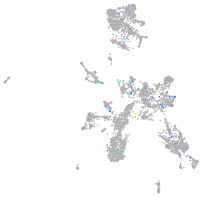
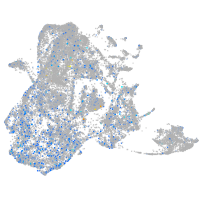
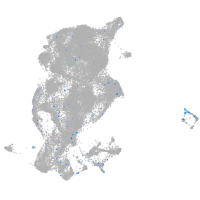
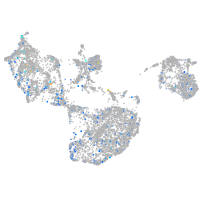
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdh6 | 0.154 | rpl37 | -0.092 |
| akap12b | 0.149 | nme2b.1 | -0.085 |
| cx43.4 | 0.148 | eef1da | -0.083 |
| pltp | 0.147 | ahcy | -0.083 |
| hspb1 | 0.146 | rps10 | -0.081 |
| prtfdc1 | 0.146 | gamt | -0.080 |
| CABZ01075068.1 | 0.138 | zgc:114188 | -0.079 |
| si:ch211-152c2.3 | 0.136 | zgc:158463 | -0.076 |
| hdac1 | 0.135 | gapdh | -0.075 |
| qkia | 0.132 | sod1 | -0.074 |
| ctnnb1 | 0.131 | gatm | -0.073 |
| id3 | 0.130 | rps17 | -0.072 |
| snrnp70 | 0.130 | nupr1b | -0.070 |
| marcksl1b | 0.129 | eno3 | -0.069 |
| khdrbs1a | 0.128 | bhmt | -0.066 |
| sfrp1a | 0.127 | pklr | -0.064 |
| tp53inp2 | 0.127 | zgc:92744 | -0.063 |
| hmga1a | 0.126 | mat1a | -0.061 |
| ppig | 0.126 | pvalb2 | -0.061 |
| zmat2 | 0.126 | gstp1 | -0.061 |
| zgc:110425 | 0.126 | atp5if1b | -0.060 |
| nucks1a | 0.126 | pvalb1 | -0.060 |
| syncrip | 0.125 | suclg1 | -0.059 |
| smc1al | 0.125 | agxtb | -0.058 |
| asph | 0.125 | gstt1a | -0.058 |
| hapln1b | 0.125 | atp5mf | -0.057 |
| seta | 0.124 | aldob | -0.057 |
| hnrnpaba | 0.123 | prdx2 | -0.057 |
| marcksb | 0.123 | apoa1b | -0.057 |
| srrm2 | 0.123 | BX908782.3 | -0.057 |
| ilf3b | 0.122 | apoa2 | -0.056 |
| LOC103910947 | 0.122 | gnmt | -0.056 |
| acin1a | 0.122 | rpl22l1 | -0.056 |
| hnrnpa1a | 0.121 | sod2 | -0.055 |
| rtf1 | 0.121 | actc1b | -0.055 |