"finTRIM family, member 82"
ZFIN 
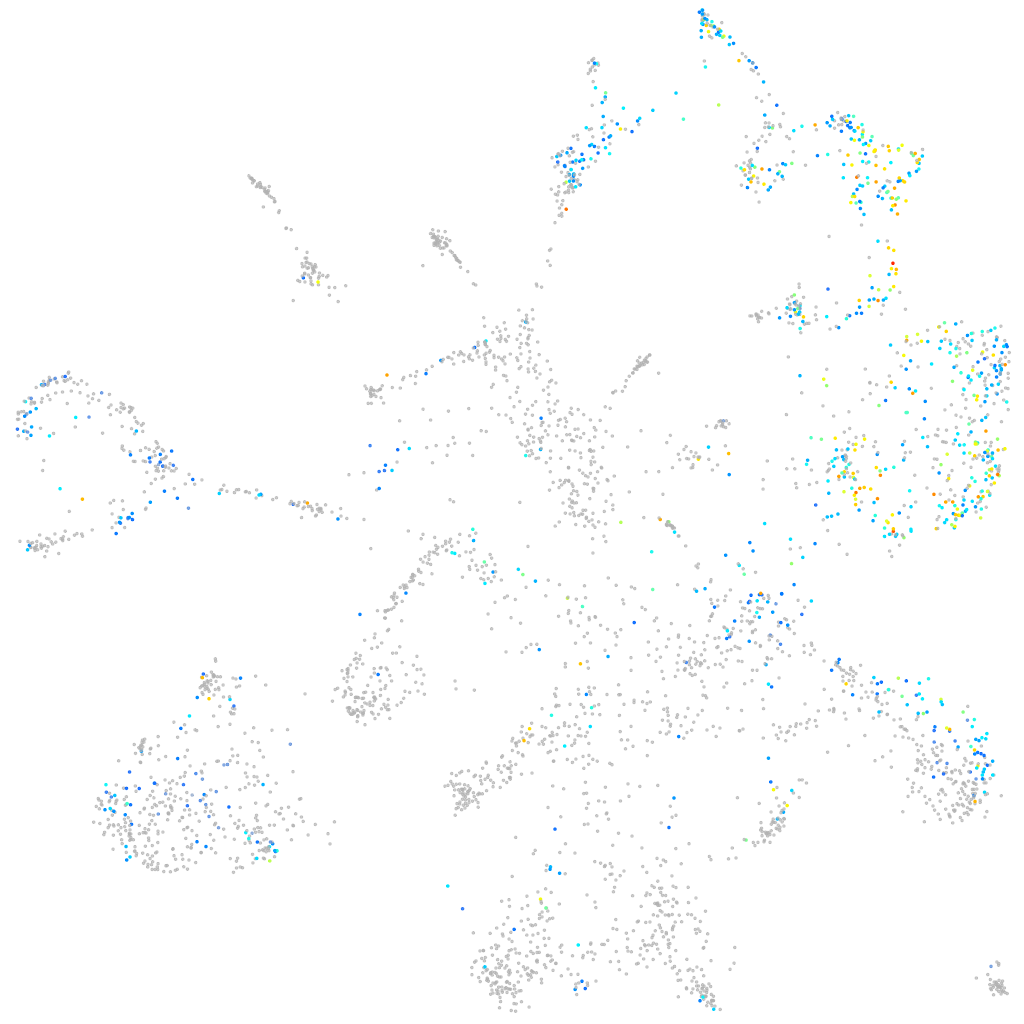
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| jam2b | 0.524 | BX088707.3 | -0.306 |
| podxl | 0.505 | tpm1 | -0.254 |
| tmem88b | 0.472 | tagln | -0.231 |
| krt94 | 0.453 | acta2 | -0.227 |
| cd151 | 0.452 | myh11a | -0.219 |
| cavin2b | 0.390 | mylkb | -0.206 |
| krt8 | 0.384 | myl9a | -0.204 |
| bnc2 | 0.356 | inka1a | -0.202 |
| COLEC10 | 0.346 | ptmaa | -0.197 |
| cav1 | 0.343 | ckbb | -0.189 |
| gata5 | 0.340 | tpm2 | -0.177 |
| si:ch211-156j16.1 | 0.330 | lmod1b | -0.175 |
| bcam | 0.328 | desmb | -0.171 |
| cavin1b | 0.327 | csrp1b | -0.167 |
| krt18a.1 | 0.326 | cald1b | -0.159 |
| tgm2b | 0.326 | XLOC-025423 | -0.158 |
| fabp11a | 0.326 | pdgfrb | -0.157 |
| tmem88a | 0.309 | igfbp7 | -0.153 |
| aqp1a.1 | 0.308 | myl6 | -0.153 |
| scarb2a | 0.307 | fhl2a | -0.152 |
| cx43.4 | 0.305 | cnn1b | -0.150 |
| tjp2b | 0.303 | gapdhs | -0.149 |
| tmem98 | 0.299 | tmem119b | -0.141 |
| hspb1 | 0.295 | si:ch211-62a1.3 | -0.140 |
| cdon | 0.294 | lbh | -0.139 |
| gata6 | 0.294 | sat1a.2 | -0.138 |
| phactr4a | 0.294 | BX323087.1 | -0.138 |
| crb2a | 0.289 | nkx2.3 | -0.137 |
| sfrp5 | 0.286 | myocd | -0.136 |
| akap12b | 0.283 | gng2 | -0.134 |
| ehd1b | 0.282 | cox4i2 | -0.133 |
| col12a1a | 0.281 | mdkb | -0.130 |
| tnni1b | 0.280 | foxl1 | -0.129 |
| gstm.3 | 0.279 | hic1 | -0.129 |
| metrnlb | 0.278 | kcne4 | -0.129 |