"ferritin, heavy polypeptide-like 29"
ZFIN 
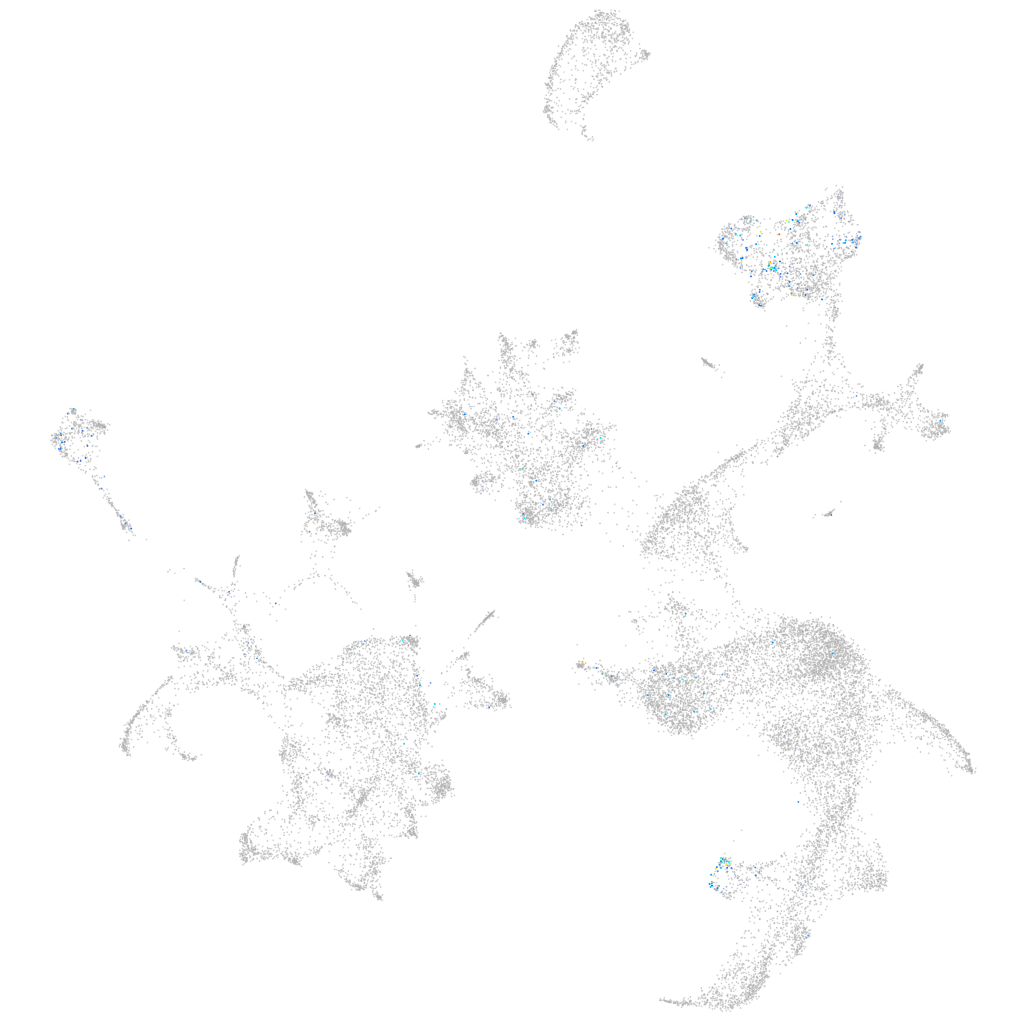
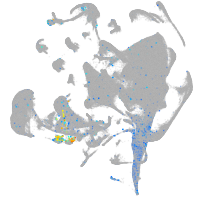
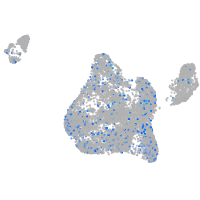
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fthl28 | 0.399 | lmo2 | -0.038 |
| fthl30 | 0.230 | si:dkey-261h17.1 | -0.038 |
| hmox1a | 0.222 | si:ch211-250g4.3 | -0.038 |
| BX511021.1 | 0.216 | krt18a.1 | -0.035 |
| fthl31 | 0.197 | tpm4a | -0.035 |
| trpv6 | 0.182 | hbbe2 | -0.034 |
| FP016234.1 | 0.181 | tmod4 | -0.033 |
| illr3 | 0.157 | hemgn | -0.033 |
| zgc:64065 | 0.155 | fli1a | -0.032 |
| lgals2a | 0.155 | TCIM (1 of many) | -0.032 |
| marco | 0.153 | creg1 | -0.031 |
| LOC100333926 | 0.150 | si:ch211-156j16.1 | -0.031 |
| fthl27 | 0.149 | cdh5 | -0.031 |
| NPC2 (1 of many) | 0.142 | aqp1a.1 | -0.031 |
| hsd17b14 | 0.134 | cldn5b | -0.030 |
| BX284666.2 | 0.132 | tuba8l3 | -0.030 |
| ctsc | 0.130 | cald1a | -0.030 |
| cndp2 | 0.127 | she | -0.030 |
| ctsl.1 | 0.126 | hbae3 | -0.029 |
| si:dkey-5n18.1 | 0.125 | zgc:56095 | -0.029 |
| havcr1 | 0.122 | ramp2 | -0.029 |
| si:ch211-283g2.2 | 0.122 | cahz | -0.029 |
| ctsba | 0.122 | hbae1.1 | -0.029 |
| pon1 | 0.122 | clec14a | -0.028 |
| mpp1 | 0.122 | myct1a | -0.028 |
| paox1 | 0.119 | tmem88a | -0.028 |
| si:dkey-82k12.13 | 0.118 | dap1b | -0.028 |
| ctsk | 0.117 | myl9a | -0.028 |
| gpd1a | 0.116 | plac8l1 | -0.028 |
| psap | 0.115 | egfl7 | -0.028 |
| zgc:174904 | 0.115 | crip2 | -0.027 |
| si:ch211-194m7.3 | 0.113 | histh1l | -0.027 |
| s100a11 | 0.113 | col4a1 | -0.027 |
| aifm4 | 0.113 | ecscr | -0.027 |
| sgk2b | 0.113 | ptprga | -0.027 |