"ferritin, heavy polypeptide-like 28"
ZFIN 
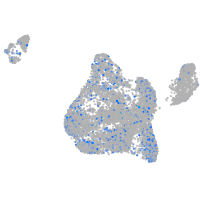
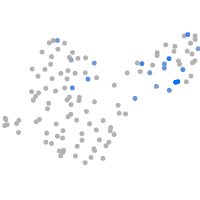
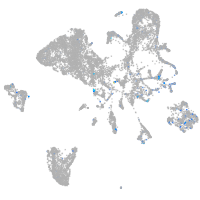
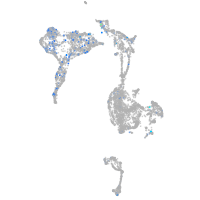
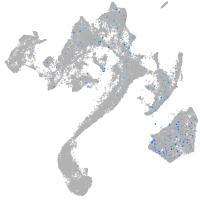
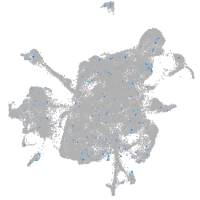
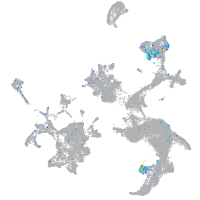
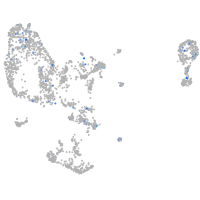
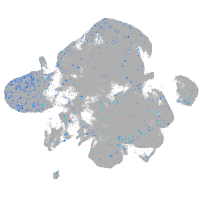
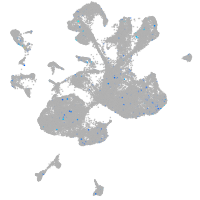
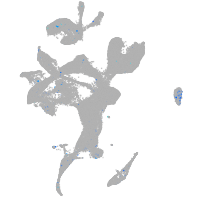
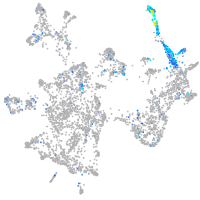
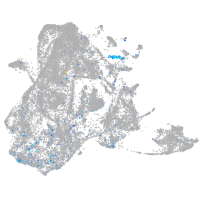
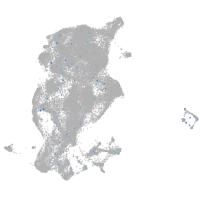
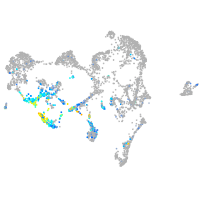
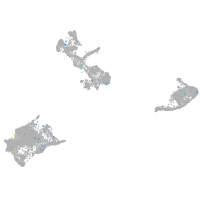
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fthl29 | 0.399 | si:ch211-250g4.3 | -0.067 |
| BX511021.1 | 0.328 | tmod4 | -0.060 |
| ctsl.1 | 0.317 | hbbe2 | -0.058 |
| fthl31 | 0.314 | krt18a.1 | -0.057 |
| fthl30 | 0.301 | creg1 | -0.057 |
| marco | 0.286 | si:dkey-261h17.1 | -0.057 |
| hmox1a | 0.272 | cdh5 | -0.053 |
| fabp7b | 0.263 | jpt1b | -0.052 |
| fthl27 | 0.263 | si:ch211-156j16.1 | -0.052 |
| NPC2 (1 of many) | 0.262 | hemgn | -0.052 |
| si:dkey-5n18.1 | 0.260 | clec14a | -0.051 |
| si:ch211-147m6.1 | 0.258 | tpm4a | -0.051 |
| havcr1 | 0.256 | aqp1a.1 | -0.050 |
| si:ch211-194m7.3 | 0.255 | crip2 | -0.050 |
| ctsba | 0.252 | hbae1.1 | -0.049 |
| ctsc | 0.249 | col4a1 | -0.048 |
| cmklr1 | 0.248 | plac8l1 | -0.048 |
| mpeg1.1 | 0.248 | lmo2 | -0.048 |
| ctss2.2 | 0.247 | she | -0.048 |
| me1 | 0.238 | zgc:56095 | -0.047 |
| zgc:174904 | 0.238 | ecscr | -0.047 |
| lgals2a | 0.237 | qkia | -0.046 |
| grna | 0.233 | cahz | -0.046 |
| c1qc | 0.232 | si:dkeyp-97a10.2 | -0.046 |
| slc7a7 | 0.230 | cldn5b | -0.046 |
| stoml3b | 0.227 | ramp2 | -0.046 |
| mfap4 | 0.224 | tuba8l3 | -0.046 |
| si:ch211-212k18.7 | 0.223 | macf1a | -0.046 |
| ctsk | 0.223 | TCIM (1 of many) | -0.046 |
| cxcl19 | 0.222 | egfl7 | -0.046 |
| ccl35.1 | 0.222 | fam174b | -0.045 |
| psap | 0.222 | igfbp7 | -0.045 |
| si:ch211-283g2.2 | 0.220 | akap12b | -0.045 |
| dnase2 | 0.220 | ptprga | -0.045 |
| lygl1 | 0.218 | tmem88a | -0.045 |