forkhead box P2
ZFIN 
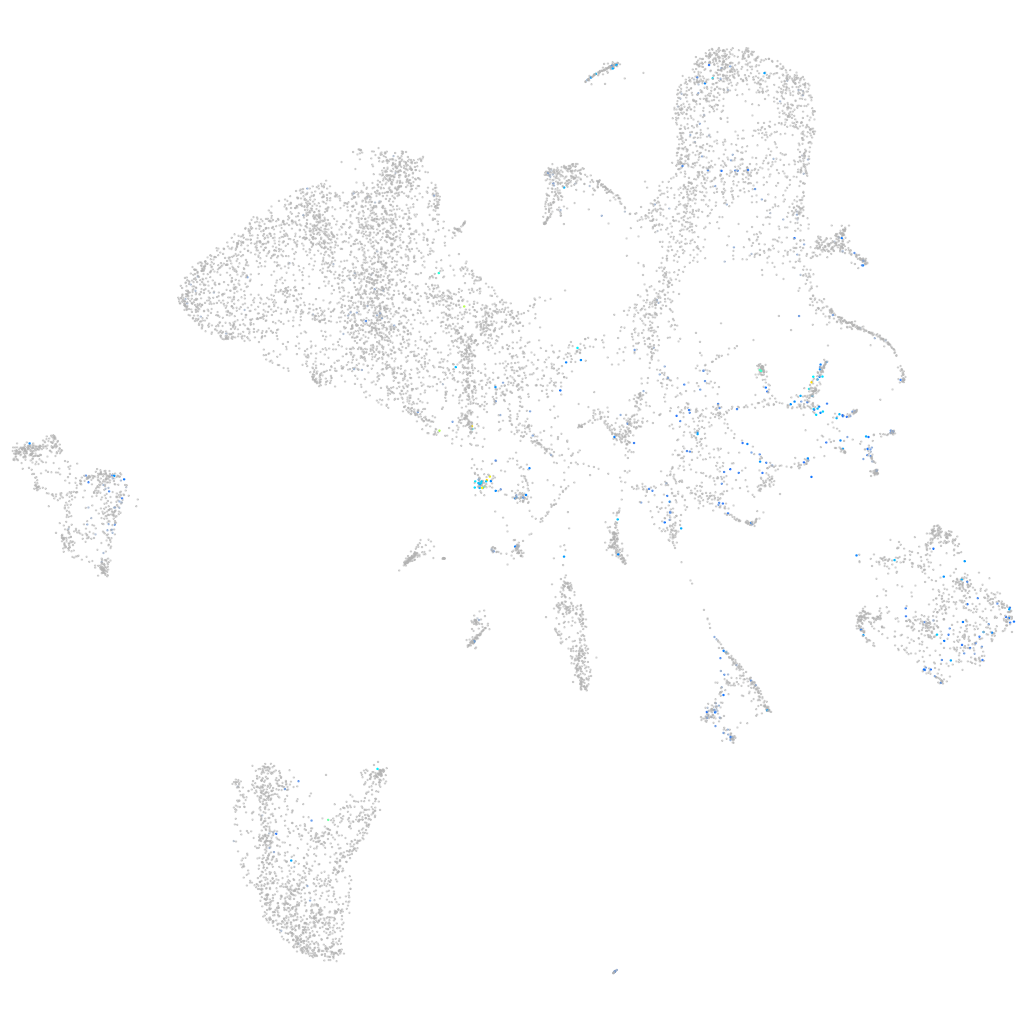
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| myt1la | 0.212 | gapdh | -0.129 |
| gpm6aa | 0.203 | ahcy | -0.119 |
| nova2 | 0.197 | gamt | -0.115 |
| stmn1b | 0.196 | gatm | -0.112 |
| fam131bb | 0.191 | fbp1b | -0.110 |
| dpysl2b | 0.182 | eno3 | -0.104 |
| cdk5r1b | 0.181 | mat1a | -0.100 |
| si:ch211-257p13.3 | 0.180 | gnmt | -0.098 |
| gng3 | 0.180 | suclg1 | -0.097 |
| elavl3 | 0.179 | nupr1b | -0.095 |
| FO082781.1 | 0.179 | aldh6a1 | -0.095 |
| scrt2 | 0.174 | aldob | -0.094 |
| hunk | 0.172 | agxtb | -0.094 |
| nos1apb | 0.172 | glud1b | -0.094 |
| atpv0e2 | 0.170 | scp2a | -0.093 |
| celf2 | 0.166 | ckba | -0.091 |
| rtn1b | 0.162 | bhmt | -0.091 |
| fez1 | 0.162 | agxta | -0.091 |
| csdc2a | 0.161 | pnp4b | -0.091 |
| hmgb3a | 0.161 | cx32.3 | -0.091 |
| sox4a | 0.159 | gstt1a | -0.090 |
| uncx | 0.159 | shmt1 | -0.090 |
| pcdh7a | 0.158 | apoc2 | -0.088 |
| cacnb4b | 0.154 | gpx4a | -0.088 |
| zc4h2 | 0.152 | pgm1 | -0.088 |
| emx2 | 0.152 | apoa1b | -0.088 |
| znf219 | 0.150 | tdo2a | -0.088 |
| cspg5a | 0.148 | cat | -0.087 |
| fyna | 0.148 | g6pca.2 | -0.087 |
| fam171b | 0.146 | grhprb | -0.087 |
| scgn | 0.146 | eef1da | -0.086 |
| myt1a | 0.146 | aqp12 | -0.086 |
| elavl4 | 0.146 | apoa4b.1 | -0.085 |
| si:dkey-12h9.6 | 0.146 | aldh1l1 | -0.085 |
| sncb | 0.145 | nipsnap3a | -0.085 |