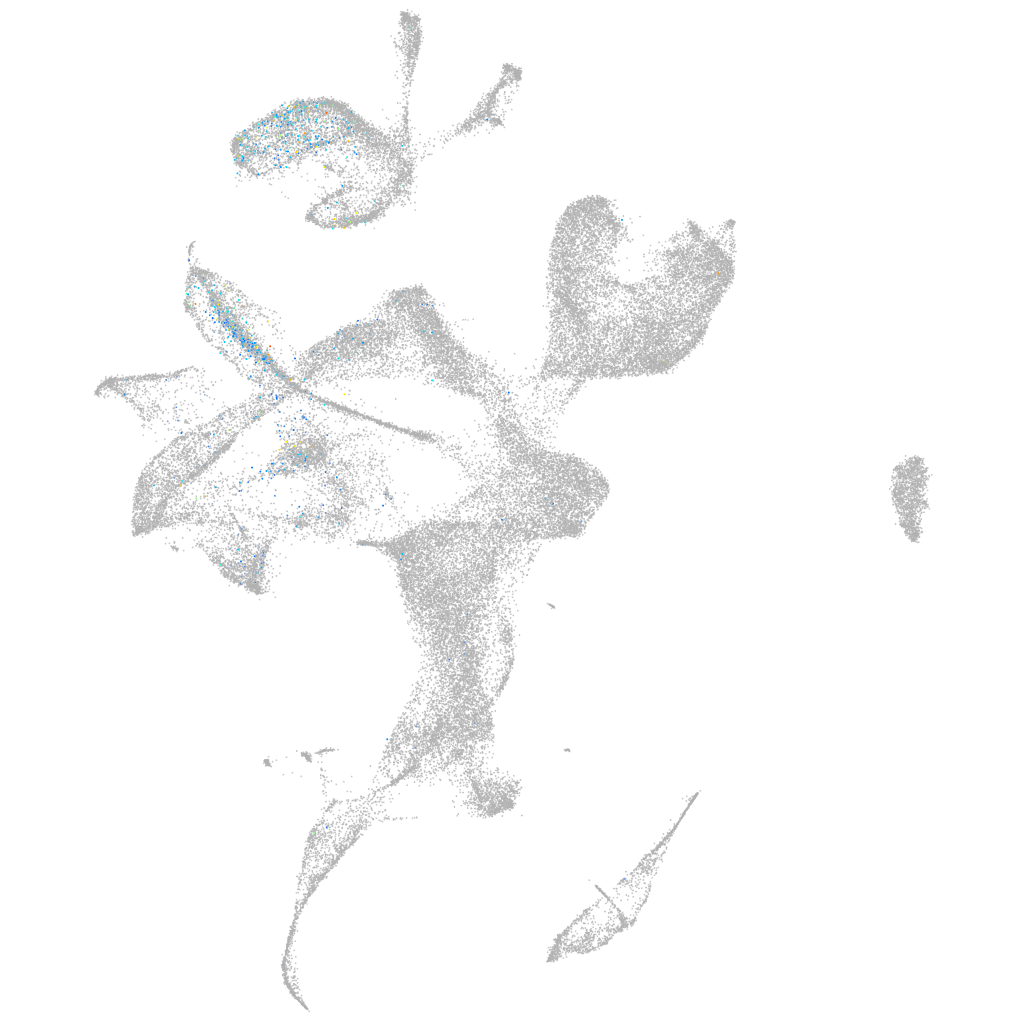
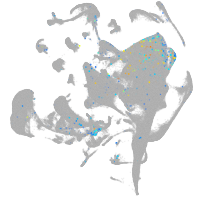
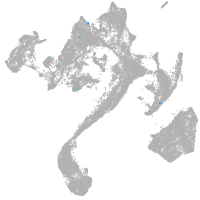
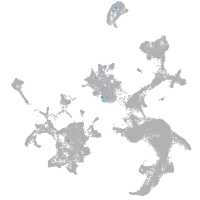
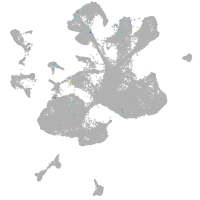
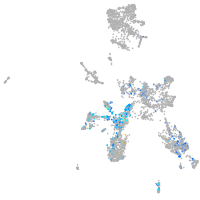
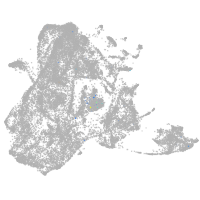
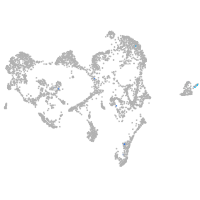
forkhead box O6 a
ZFIN 
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rtn1b | 0.154 | hmgb2a | -0.105 |
| ppp1r14ba | 0.153 | mdka | -0.058 |
| stmn2a | 0.152 | stmn1a | -0.057 |
| elavl3 | 0.147 | pcna | -0.057 |
| zgc:153426 | 0.145 | rrm1 | -0.054 |
| gng3 | 0.141 | mki67 | -0.054 |
| ywhag2 | 0.141 | tuba8l | -0.053 |
| stmn2b | 0.141 | dut | -0.053 |
| pbx1a | 0.139 | ahcy | -0.052 |
| pbx3b | 0.138 | msna | -0.052 |
| zgc:65894 | 0.138 | fabp7a | -0.052 |
| sncb | 0.137 | selenoh | -0.052 |
| pax10 | 0.137 | ccnd1 | -0.051 |
| stxbp1a | 0.136 | lbr | -0.051 |
| id4 | 0.133 | chaf1a | -0.051 |
| si:ch73-290k24.5 | 0.133 | nutf2l | -0.050 |
| eno2 | 0.132 | mcm7 | -0.050 |
| stx1b | 0.131 | ccna2 | -0.049 |
| maptb | 0.130 | banf1 | -0.047 |
| meis2b | 0.128 | COX7A2 (1 of many) | -0.047 |
| mab21l1 | 0.126 | her15.1 | -0.047 |
| pou4f2 | 0.125 | dek | -0.047 |
| gap43 | 0.125 | neurod4 | -0.046 |
| rbpms2a | 0.125 | cks1b | -0.046 |
| stmn1b | 0.124 | lig1 | -0.045 |
| LOC101886114 | 0.123 | tspan7 | -0.045 |
| tubb5 | 0.122 | nasp | -0.045 |
| si:ch211-129p13.1 | 0.120 | rpa3 | -0.044 |
| mir181b-3 | 0.120 | zgc:110425 | -0.044 |
| celf5a | 0.118 | CABZ01005379.1 | -0.044 |
| gabrb4 | 0.118 | mcm6 | -0.043 |
| myt1la | 0.118 | cdca7b | -0.043 |
| syt1a | 0.117 | rrm2 | -0.043 |
| gnao1a | 0.116 | mcm5 | -0.043 |
| pou4f1 | 0.116 | eef1da | -0.043 |