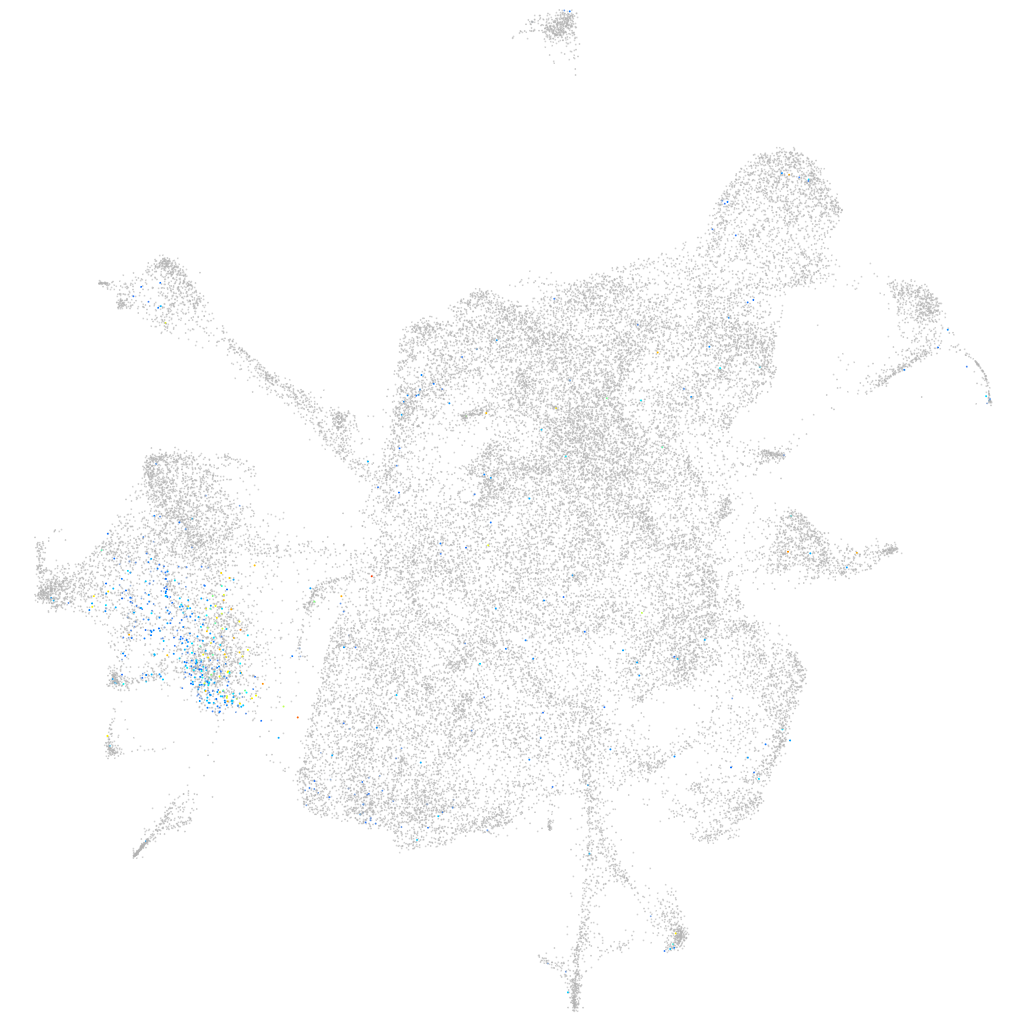
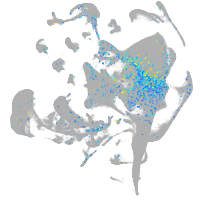
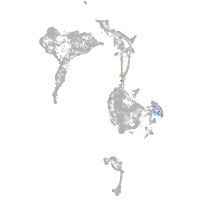
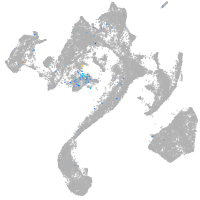
forkhead box N4
ZFIN 
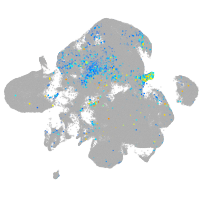
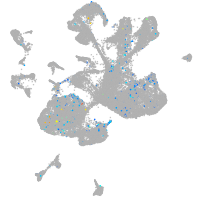
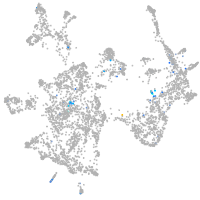
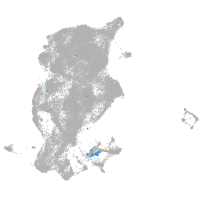
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hes2.2 | 0.310 | pmp22a | -0.065 |
| hes2.1 | 0.228 | sparc | -0.059 |
| insm1a | 0.224 | mfap2 | -0.052 |
| neurod4 | 0.179 | krt8 | -0.050 |
| cadm3 | 0.177 | krt18a.1 | -0.046 |
| fabp7a | 0.175 | tpm4a | -0.045 |
| hes6 | 0.172 | eef1da | -0.040 |
| im:7152348 | 0.166 | bhmt | -0.038 |
| rorb | 0.156 | krt18b | -0.037 |
| si:ch211-137a8.4 | 0.155 | cd63 | -0.036 |
| otx2b | 0.154 | fstl1b | -0.036 |
| dla | 0.152 | sdc4 | -0.035 |
| gadd45gb.1 | 0.151 | fkbp7 | -0.035 |
| CU634008.1 | 0.149 | col5a1 | -0.035 |
| atoh7 | 0.144 | twist1a | -0.034 |
| prdm1b | 0.144 | si:ch1073-291c23.2 | -0.034 |
| her15.1 | 0.131 | xbp1 | -0.033 |
| msi1 | 0.130 | ppib | -0.033 |
| XLOC-003692 | 0.127 | col2a1b | -0.033 |
| CU467822.1 | 0.127 | col1a2 | -0.032 |
| XLOC-003690 | 0.125 | zgc:162730 | -0.032 |
| olig2 | 0.125 | aldh9a1a.1 | -0.032 |
| foxg1b | 0.124 | tmem176 | -0.031 |
| notch1a | 0.123 | ms4a17a.9 | -0.031 |
| tuba1a | 0.122 | mmp2 | -0.031 |
| sept4a | 0.122 | anxa11b | -0.031 |
| ndrg1b | 0.112 | zgc:153675 | -0.031 |
| abhd6a | 0.112 | foxc1b | -0.031 |
| sox3 | 0.111 | si:ch211-286o17.1 | -0.030 |
| her4.2 | 0.105 | fmoda | -0.030 |
| mki67 | 0.105 | ckap4 | -0.030 |
| gpm6aa | 0.104 | zgc:153867 | -0.030 |
| XLOC-014477 | 0.103 | ier2b | -0.030 |
| nusap1 | 0.102 | crtap | -0.030 |
| LOC798783 | 0.101 | rarga | -0.029 |