forkhead box I3b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line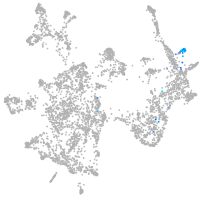 |
epidermis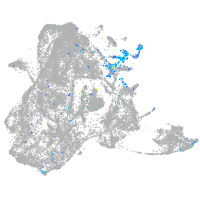 |
periderm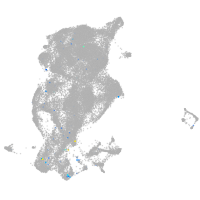 |
fin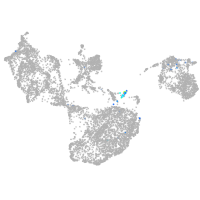 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-33i11.4 | 0.685 | mfap2 | -0.037 |
| sstr5 | 0.582 | hmga2 | -0.034 |
| foxi3a | 0.564 | marcksl1a | -0.033 |
| atp1b1b | 0.504 | mxra8b | -0.032 |
| mal2 | 0.416 | lrrc17 | -0.031 |
| atp1a1a.2 | 0.371 | crabp2a | -0.031 |
| myb | 0.356 | bhmt | -0.031 |
| ceacam1 | 0.352 | twist1a | -0.030 |
| gcm2 | 0.349 | c1qtnf5 | -0.030 |
| LOC799574 | 0.348 | tmem119b | -0.030 |
| si:dkeyp-97e7.9 | 0.318 | rpl31 | -0.030 |
| kcnj1a.5 | 0.317 | prrx1b | -0.029 |
| cldnh | 0.316 | hoxc13a | -0.029 |
| kcnj1a.6 | 0.302 | prrx1a | -0.029 |
| CU915778.1 | 0.302 | cd82a | -0.029 |
| LOC101884089 | 0.302 | dcn | -0.029 |
| CR356236.1 | 0.300 | ntd5 | -0.028 |
| ca15a | 0.297 | pmp22a | -0.028 |
| kcnj1a.2 | 0.293 | boc | -0.028 |
| CR927456.1 | 0.290 | stox1 | -0.028 |
| esrrgb | 0.290 | cd81a | -0.027 |
| nipal4 | 0.290 | sept2 | -0.027 |
| slc16a12a | 0.290 | pcbd1 | -0.027 |
| trim35-12 | 0.282 | ptx3a | -0.027 |
| ndrg1a | 0.279 | reck | -0.027 |
| si:dkey-192d15.2 | 0.265 | hoxa13b | -0.026 |
| CU914776.2 | 0.265 | MDFI | -0.026 |
| si:ch73-359m17.5 | 0.264 | zgc:153675 | -0.026 |
| si:ch211-270g19.5 | 0.262 | fbln1 | -0.026 |
| BX640453.1 | 0.242 | hgd | -0.026 |
| BX465848.1 | 0.237 | crabp1b | -0.025 |
| slc4a1b | 0.237 | nck1a | -0.025 |
| sgk2a | 0.229 | si:ch211-133n4.4 | -0.025 |
| slc12a10.2 | 0.226 | pdgfra | -0.025 |
| si:ch73-31d8.2 | 0.225 | hoxa13a | -0.025 |




