FKBP prolyl isomerase 14
ZFIN 
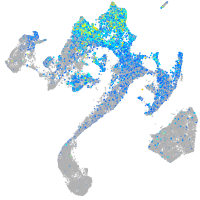
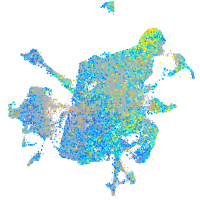
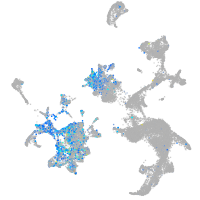
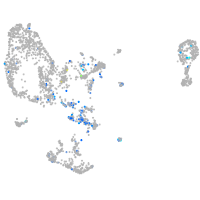
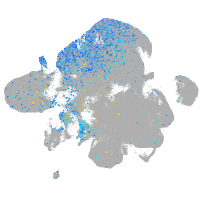
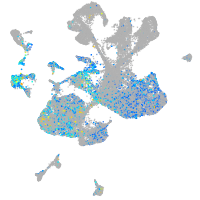
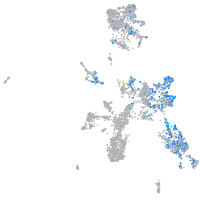
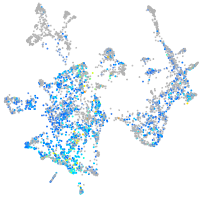
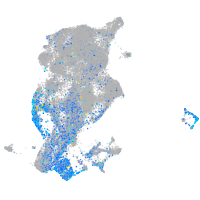
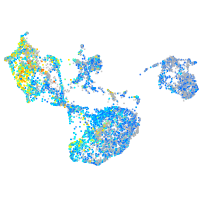
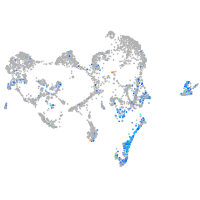
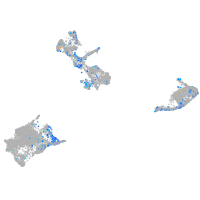
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cnn2 | 0.365 | cldnh | -0.181 |
| krt18b | 0.360 | tuba1c | -0.180 |
| si:ch211-243g18.2 | 0.346 | calm1a | -0.168 |
| her6 | 0.330 | vamp2 | -0.165 |
| sparc | 0.323 | rnasekb | -0.164 |
| krt8 | 0.311 | rtn1a | -0.164 |
| sp8a | 0.292 | scg2a | -0.152 |
| cfl1l | 0.286 | atp6v0cb | -0.151 |
| id1 | 0.286 | txn | -0.150 |
| six1b | 0.286 | insm1a | -0.149 |
| dmrt3a | 0.284 | elavl3 | -0.146 |
| zfp36l1a | 0.280 | anxa5b | -0.136 |
| tpm4a | 0.274 | atp6v1e1b | -0.135 |
| tmsb4x | 0.273 | stmn1b | -0.135 |
| junba | 0.273 | COX3 | -0.125 |
| zgc:158343 | 0.271 | nfasca | -0.124 |
| fxyd6l | 0.271 | epb41a | -0.120 |
| rac1a | 0.270 | gng3 | -0.119 |
| dlx5a | 0.267 | gdi1 | -0.119 |
| myh9a | 0.259 | si:dkeyp-75h12.5 | -0.119 |
| dnase1l4.1 | 0.259 | kcnc1a | -0.119 |
| gpt2l | 0.258 | zgc:65894 | -0.118 |
| cebpb | 0.258 | atp6v1g1 | -0.118 |
| sept2 | 0.257 | ebf3a | -0.117 |
| krt97 | 0.257 | bik | -0.116 |
| s100a10b | 0.256 | sncb | -0.116 |
| crtap | 0.255 | atp2b1b | -0.115 |
| six3a | 0.253 | tspan2a | -0.115 |
| spaca4l | 0.251 | insm1b | -0.115 |
| si:dkeyp-86h10.3 | 0.251 | eml2 | -0.114 |
| lgals3b | 0.250 | zgc:165573 | -0.114 |
| si:dkey-261h17.1 | 0.250 | calm1b | -0.113 |
| col18a1a | 0.249 | chga | -0.112 |
| pak2a | 0.245 | si:ch73-199e17.1 | -0.112 |
| XLOC-044191 | 0.245 | bcl2l10 | -0.112 |