fin bud initiation factor a
ZFIN 
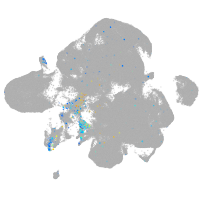
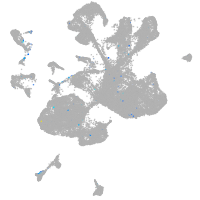
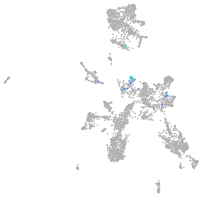
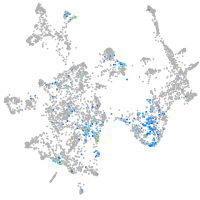
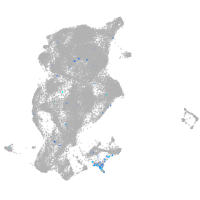
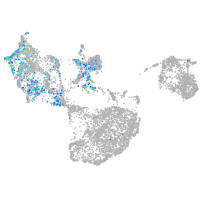
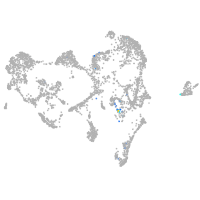
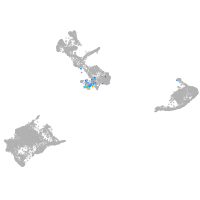
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| col11a1b | 0.379 | impdh1b | -0.074 |
| mfap2 | 0.371 | tspan36 | -0.067 |
| col5a1 | 0.362 | syngr1a | -0.066 |
| LO018188.1 | 0.357 | akr1b1 | -0.063 |
| foxc1b | 0.344 | paics | -0.062 |
| fmoda | 0.339 | gpr143 | -0.060 |
| col2a1b | 0.339 | gmps | -0.059 |
| six1a | 0.334 | pts | -0.059 |
| matn4 | 0.333 | slc2a15a | -0.058 |
| cd248b | 0.329 | gch2 | -0.057 |
| mxra8b | 0.328 | rab34b | -0.057 |
| col1a2 | 0.323 | rab32a | -0.056 |
| colec12 | 0.322 | mdh1aa | -0.056 |
| twist1a | 0.320 | atic | -0.056 |
| lrrc17 | 0.316 | slc2a11b | -0.056 |
| dcn | 0.315 | trpm1b | -0.055 |
| foxc1a | 0.304 | glulb | -0.054 |
| tnn | 0.303 | uraha | -0.054 |
| si:ch1073-459j12.1 | 0.303 | rab38 | -0.053 |
| col12a1a | 0.301 | agtrap | -0.053 |
| mkxa | 0.298 | pfn1 | -0.053 |
| wfdc2 | 0.295 | gstp1 | -0.052 |
| cthrc1a | 0.292 | prdx1 | -0.052 |
| ms4a17a.9 | 0.290 | opn5 | -0.051 |
| mmp2 | 0.288 | pttg1ipb | -0.051 |
| col12a1b | 0.288 | LOC103910009 | -0.051 |
| col1a1b | 0.285 | si:dkey-21a6.5 | -0.050 |
| col9a1a | 0.285 | eno3 | -0.050 |
| fam198a | 0.284 | bace2 | -0.050 |
| pdgfrl | 0.283 | C12orf75 | -0.050 |
| CU929237.1 | 0.283 | smim29 | -0.049 |
| pmp22a | 0.279 | XLOC-019062 | -0.049 |
| olfml3b | 0.273 | atp11a | -0.049 |
| mir214a | 0.271 | slc22a7a | -0.048 |
| pdgfra | 0.271 | abracl | -0.048 |