fibroblast growth factor receptor 1a
ZFIN 
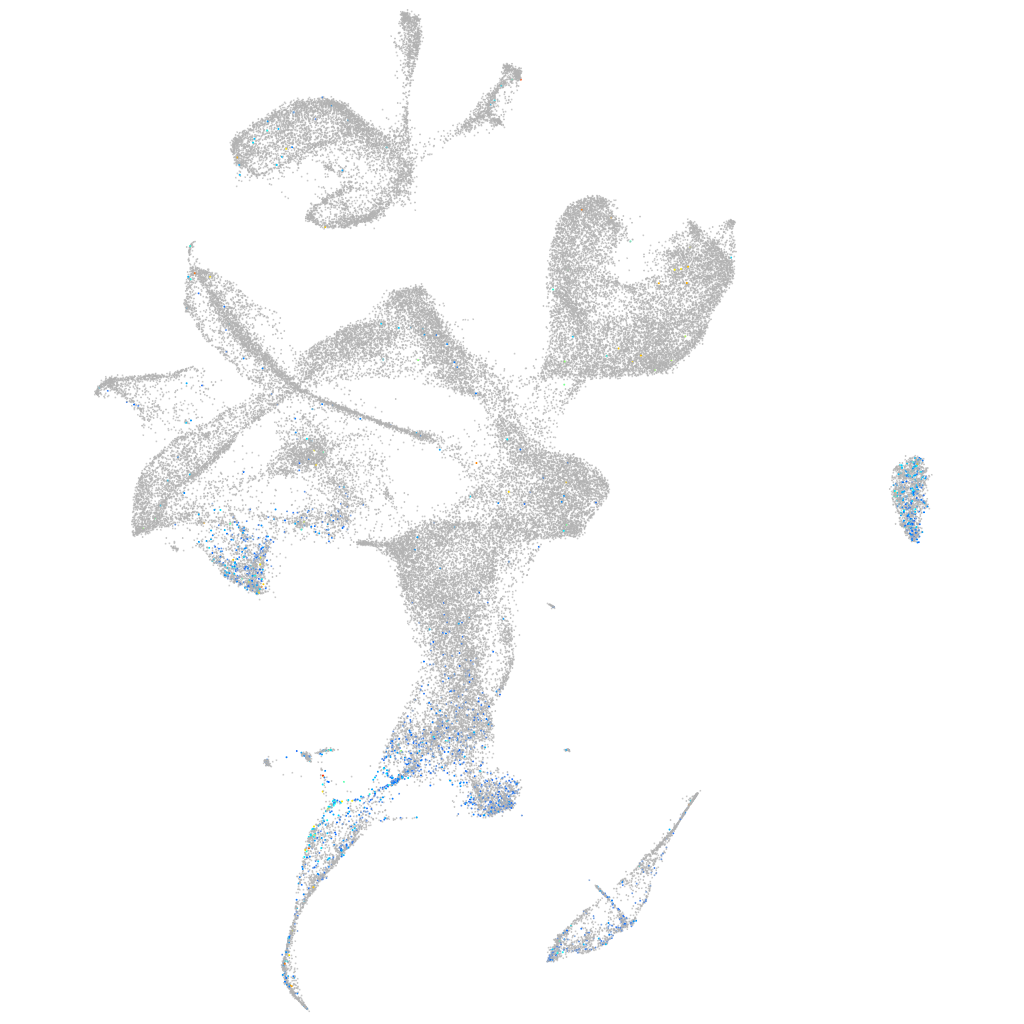
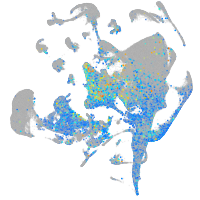
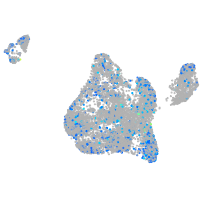
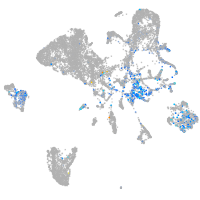
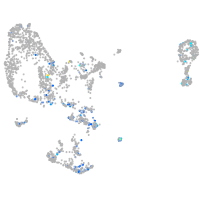
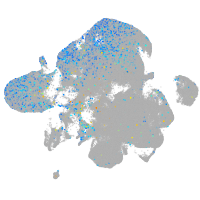
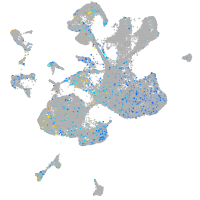
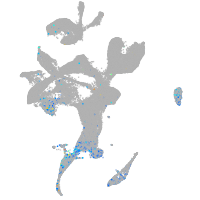
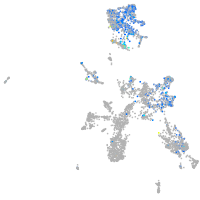
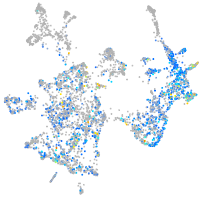
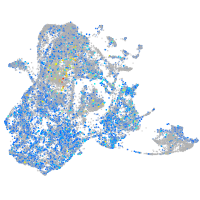
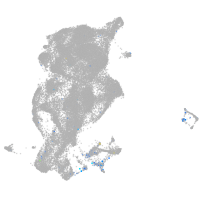
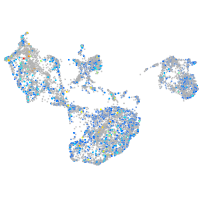
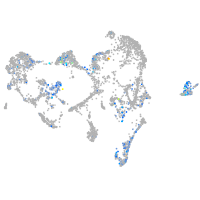
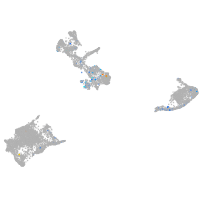
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| wls | 0.173 | ptmab | -0.121 |
| fstl1b | 0.167 | ptmaa | -0.117 |
| fabp11a | 0.166 | tuba1c | -0.107 |
| cdon | 0.161 | gpm6aa | -0.106 |
| atp1b1a | 0.159 | ckbb | -0.105 |
| tpm4a | 0.153 | cadm3 | -0.095 |
| cx43 | 0.152 | gpm6ab | -0.093 |
| sfrp1a | 0.150 | hmgn6 | -0.084 |
| hspb1 | 0.149 | fabp7a | -0.084 |
| sparc | 0.147 | foxg1b | -0.081 |
| ldlrap1a | 0.147 | h3f3a | -0.080 |
| foxp4 | 0.146 | hmgb1a | -0.078 |
| aldob | 0.144 | epb41a | -0.077 |
| tspan36 | 0.143 | rorb | -0.077 |
| lamb1a | 0.142 | tuba1a | -0.076 |
| gpc4 | 0.139 | si:ch1073-429i10.3.1 | -0.076 |
| col15a1b | 0.138 | rnasekb | -0.076 |
| fgfrl1a | 0.137 | snap25b | -0.073 |
| npm1a | 0.137 | marcksl1b | -0.072 |
| crtap | 0.136 | ppiab | -0.071 |
| slc38a5b | 0.136 | neurod4 | -0.071 |
| plxdc2 | 0.136 | btg1 | -0.069 |
| c1qtnf12 | 0.136 | crx | -0.068 |
| notch2 | 0.135 | h3f3c | -0.067 |
| otx1 | 0.135 | atp6v0cb | -0.065 |
| sdc4 | 0.135 | stmn1b | -0.064 |
| otx2a | 0.134 | ubc | -0.064 |
| boc | 0.133 | h2afva | -0.063 |
| eif4ebp3l | 0.132 | marcksl1a | -0.063 |
| hspg2 | 0.132 | ndrg4 | -0.063 |
| bzw1b | 0.132 | scrt2 | -0.063 |
| pprc1 | 0.131 | si:ch73-1a9.3 | -0.063 |
| cldn19 | 0.131 | pdcl | -0.062 |
| stm | 0.131 | syt5b | -0.062 |
| mfap2 | 0.131 | pvalb1 | -0.061 |