fibrillin 2b
ZFIN 
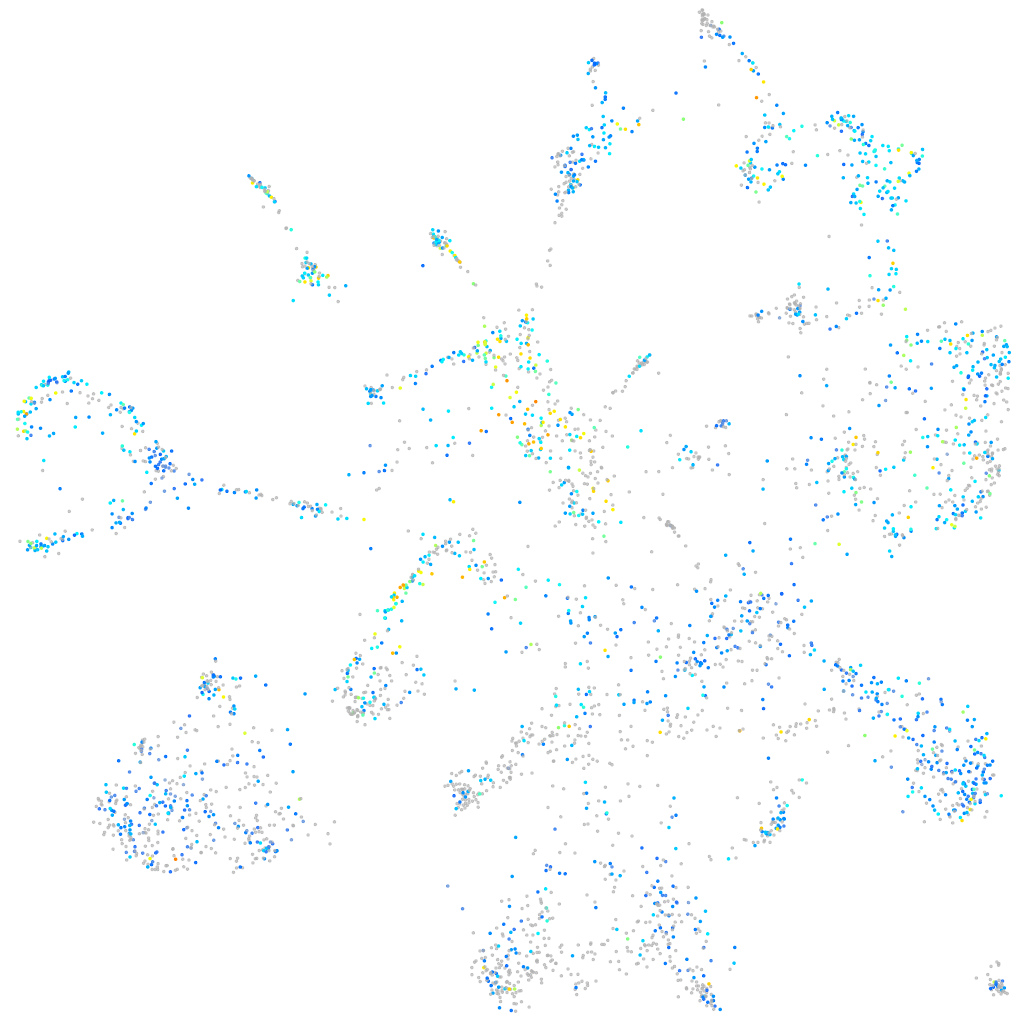
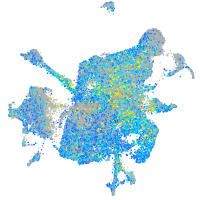
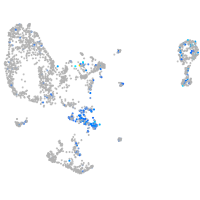
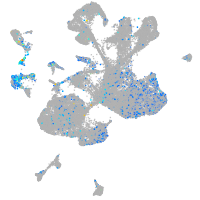
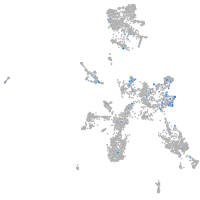
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fstl1b | 0.250 | XLOC-041870 | -0.186 |
| col1a1a | 0.225 | desmb | -0.176 |
| col1a2 | 0.223 | XLOC-025423 | -0.171 |
| akap12b | 0.208 | ckbb | -0.160 |
| ednraa | 0.202 | cfd | -0.148 |
| myh10 | 0.199 | csrp1b | -0.134 |
| col2a1b | 0.195 | rgs2 | -0.133 |
| sparc | 0.194 | rplp1 | -0.133 |
| col5a1 | 0.193 | cald1b | -0.128 |
| mfap2 | 0.182 | mdkb | -0.126 |
| col11a1b | 0.181 | si:ch211-137i24.10 | -0.123 |
| cd248a | 0.176 | mylkb | -0.118 |
| col12a1a | 0.174 | XLOC-002639 | -0.113 |
| sfrp5 | 0.173 | tpm2 | -0.112 |
| col6a4a | 0.168 | cnn1b | -0.111 |
| sdc4 | 0.167 | XLOC-028370 | -0.106 |
| cd81a | 0.164 | zgc:110699 | -0.104 |
| si:dkey-261h17.1 | 0.162 | nkx2.3 | -0.103 |
| twist1b | 0.158 | ntn5 | -0.103 |
| cald1a | 0.157 | BX322618.1 | -0.102 |
| ahnak | 0.157 | vcla | -0.102 |
| lsp1 | 0.155 | BX248318.1 | -0.101 |
| marcksl1b | 0.155 | fhl1a | -0.101 |
| hdlbpa | 0.153 | BX908750.1 | -0.100 |
| emilin2a | 0.151 | myh11a | -0.100 |
| thy1 | 0.151 | rpl37 | -0.096 |
| cnn2 | 0.150 | atp5mc3b | -0.095 |
| emid1 | 0.149 | pdlim3a | -0.095 |
| colec12 | 0.149 | acta2 | -0.092 |
| krt18a.1 | 0.148 | KCNIP4 | -0.092 |
| fbln2 | 0.148 | nptna | -0.092 |
| twist1a | 0.147 | BX005183.3 | -0.092 |
| mmel1 | 0.146 | pdlim3b | -0.091 |
| crtap | 0.146 | BX323087.1 | -0.091 |
| mfap5 | 0.146 | cabp1a | -0.091 |