fibulin 2
ZFIN 
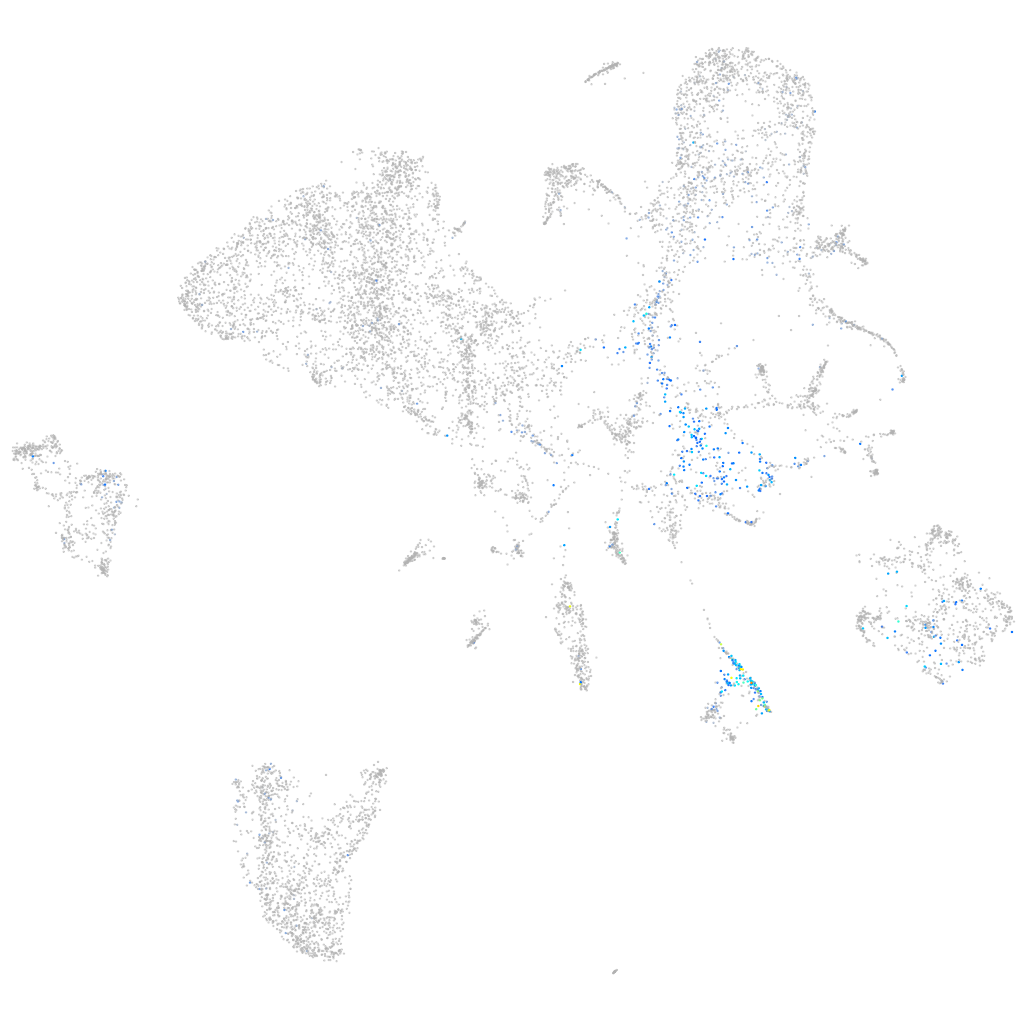
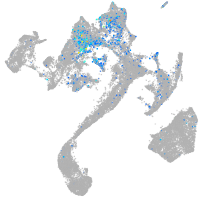
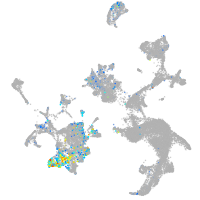
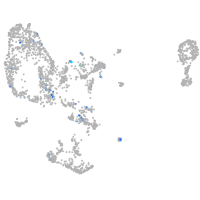
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pmp22b | 0.376 | gapdh | -0.162 |
| lama5 | 0.376 | gatm | -0.150 |
| col18a1a | 0.372 | bhmt | -0.148 |
| bcam | 0.353 | gamt | -0.145 |
| sim1b | 0.351 | mat1a | -0.137 |
| CABZ01055365.1 | 0.342 | agxtb | -0.136 |
| lamb1a | 0.337 | ahcy | -0.134 |
| ctgfa | 0.335 | gpx4a | -0.133 |
| dhrs3a | 0.332 | apoa4b.1 | -0.132 |
| tekt3 | 0.326 | pnp4b | -0.128 |
| b3gnt5a | 0.320 | fbp1b | -0.127 |
| lrata | 0.320 | apoa2 | -0.126 |
| spns2 | 0.319 | afp4 | -0.126 |
| mxra8b | 0.317 | apoa1b | -0.124 |
| anxa3b | 0.306 | sod1 | -0.124 |
| tgfb3 | 0.306 | grhprb | -0.124 |
| cd9b | 0.303 | fabp10a | -0.122 |
| agrn | 0.301 | abat | -0.122 |
| sgms1 | 0.301 | scp2a | -0.121 |
| spock3 | 0.299 | apom | -0.121 |
| tpm4a | 0.294 | zgc:123103 | -0.119 |
| abca12 | 0.292 | ces2 | -0.119 |
| myl9a | 0.291 | rbp2b | -0.118 |
| lsp1 | 0.291 | agxta | -0.118 |
| marcksl1a | 0.290 | rbp4 | -0.117 |
| f3a | 0.287 | fgb | -0.117 |
| cd151 | 0.285 | hpda | -0.117 |
| cav1 | 0.285 | kng1 | -0.116 |
| FP102018.1 | 0.283 | serpina1 | -0.116 |
| pnp4a | 0.280 | uox | -0.116 |
| arhgap29b | 0.278 | etnppl | -0.116 |
| sftpba | 0.278 | fetub | -0.115 |
| cabp2b | 0.274 | hao1 | -0.115 |
| crip2 | 0.269 | serpina1l | -0.115 |
| erbb3a | 0.268 | mdh1aa | -0.115 |