family with sequence similarity 49 member Bb
ZFIN 
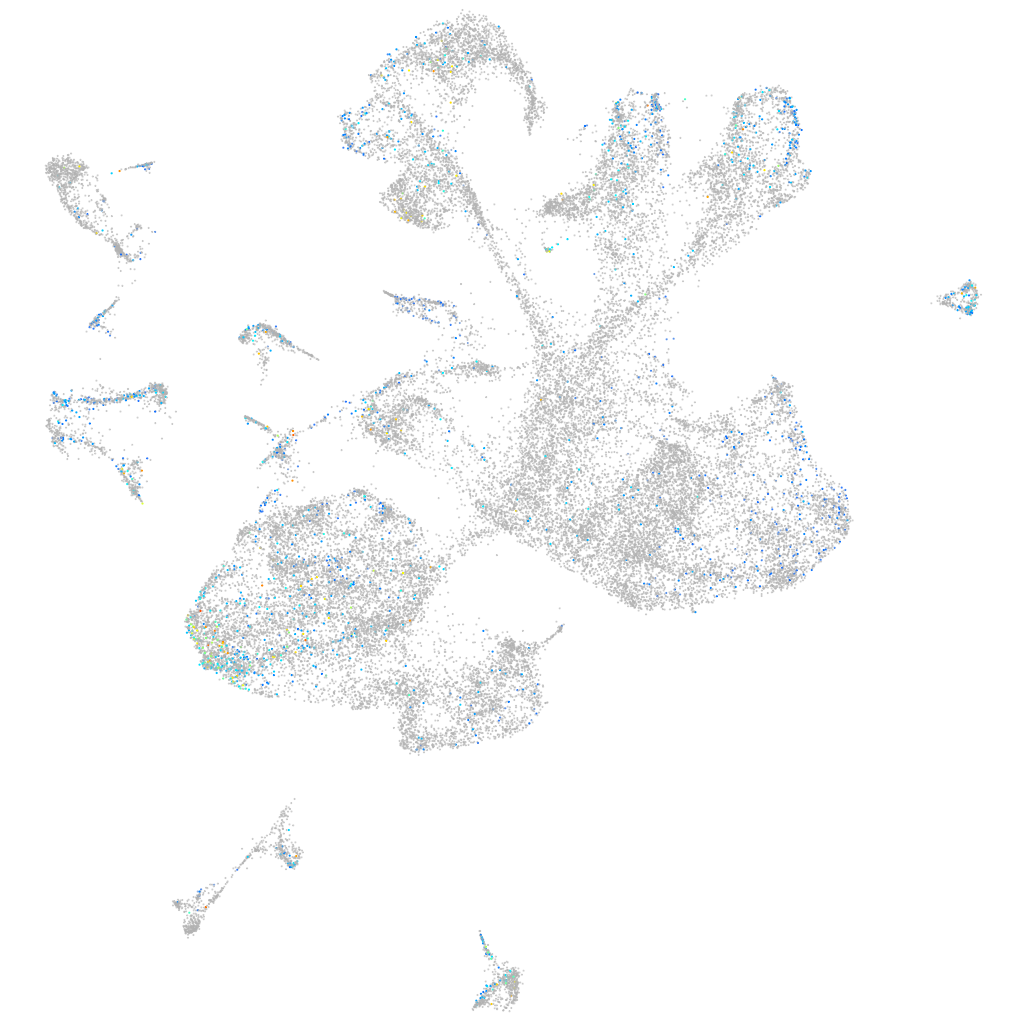
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gapdhs | 0.129 | si:ch211-222l21.1 | -0.099 |
| aldocb | 0.127 | rplp1 | -0.097 |
| ndrg3a | 0.121 | hmga1a | -0.096 |
| tpi1b | 0.116 | hmgb2b | -0.094 |
| mgll | 0.114 | si:dkey-151g10.6 | -0.092 |
| glula | 0.113 | rplp2l | -0.091 |
| anxa13 | 0.111 | rpl37 | -0.091 |
| slc6a1b | 0.110 | rps20 | -0.088 |
| mfge8a | 0.108 | rps19 | -0.086 |
| slc38a3a | 0.108 | rps12 | -0.086 |
| cx43 | 0.108 | XLOC-003689 | -0.085 |
| aqp1a.1 | 0.107 | hmgn2 | -0.085 |
| acbd7 | 0.105 | cirbpb | -0.083 |
| ckbb | 0.105 | rps21 | -0.083 |
| eno1a | 0.104 | rps14 | -0.083 |
| slc4a4a | 0.103 | hmgb2a | -0.082 |
| pkma | 0.103 | rps9 | -0.082 |
| cspg5b | 0.101 | rps25 | -0.081 |
| ptn | 0.099 | rpl36 | -0.081 |
| slc1a2b | 0.098 | hnrnpa0l | -0.079 |
| fjx1 | 0.098 | ptmab | -0.079 |
| ptgdsb.2 | 0.098 | rps15a | -0.078 |
| calm3a | 0.098 | rpl17 | -0.078 |
| sult6b1 | 0.097 | rps8a | -0.078 |
| cdo1 | 0.096 | rps5 | -0.077 |
| FO704813.1 | 0.096 | si:ch73-21g5.7 | -0.076 |
| atp6v1e1b | 0.095 | faua | -0.076 |
| pygmb | 0.094 | khdrbs1a | -0.076 |
| efhd1 | 0.093 | rps23 | -0.076 |
| calm1b | 0.092 | rpl13 | -0.076 |
| ca4a | 0.092 | rpl36a | -0.075 |
| gpia | 0.091 | rpl11 | -0.075 |
| pgk1 | 0.090 | rpl18 | -0.075 |
| slc3a2a | 0.090 | rpl23 | -0.075 |
| si:ch211-66e2.5 | 0.090 | rpl39 | -0.074 |