family with sequence similarity 171 member A2a
ZFIN 
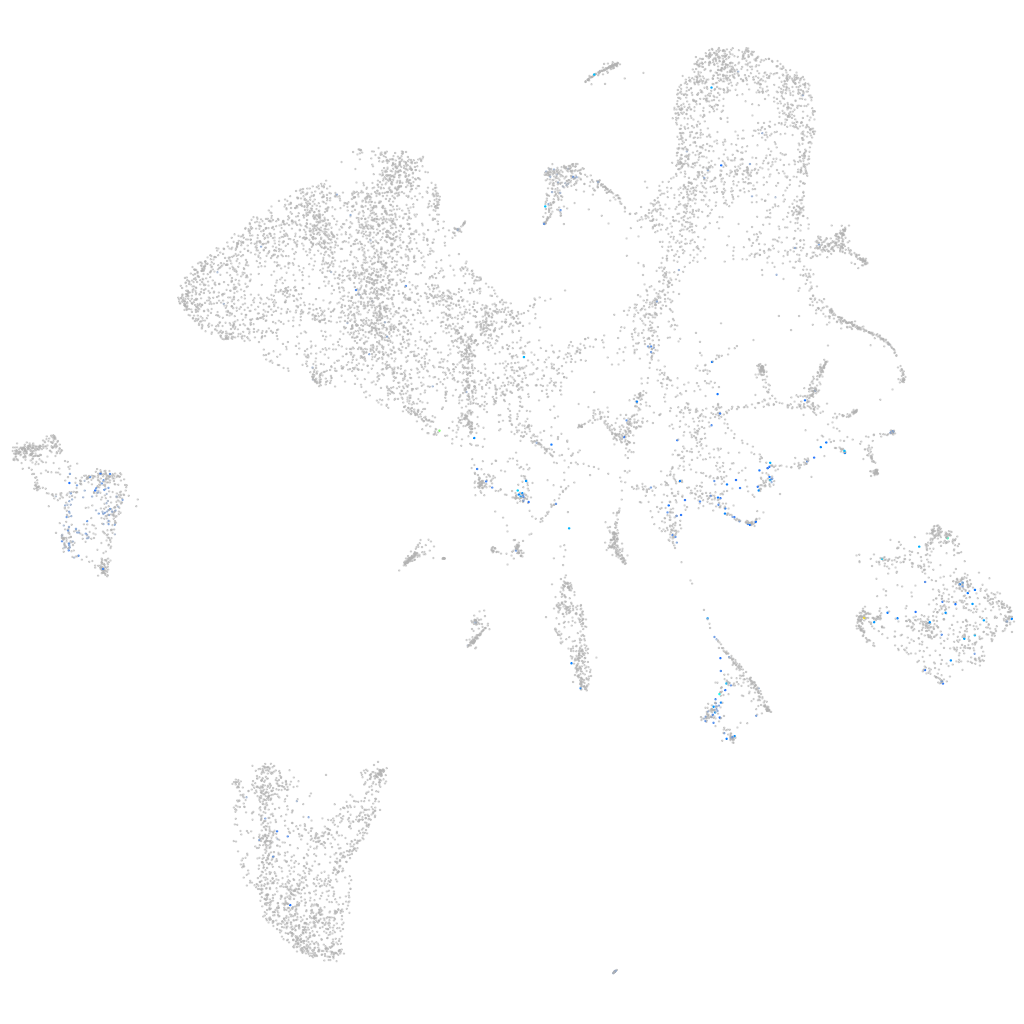
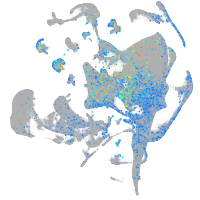
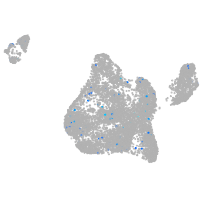
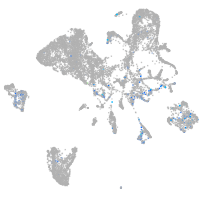
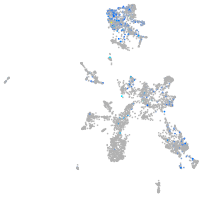
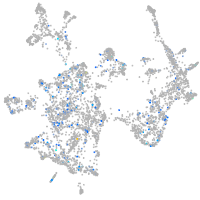
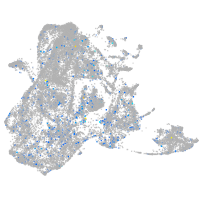
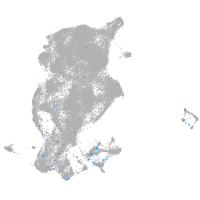
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU633479.5 | 0.166 | gapdh | -0.129 |
| il1rapl2 | 0.153 | gamt | -0.122 |
| msna | 0.142 | gatm | -0.109 |
| h3f3d | 0.138 | pklr | -0.109 |
| sox3 | 0.137 | fbp1b | -0.108 |
| cx43.4 | 0.136 | gpx4a | -0.106 |
| bcam | 0.134 | ahcy | -0.103 |
| si:ch211-222l21.1 | 0.134 | eno3 | -0.103 |
| hmgb1b | 0.132 | aldob | -0.102 |
| hmgb2a | 0.131 | mat1a | -0.101 |
| qkia | 0.130 | scp2a | -0.100 |
| fgfbp1b | 0.130 | apoa4b.1 | -0.098 |
| h2afvb | 0.129 | apoa1b | -0.098 |
| ndnf | 0.129 | apoc2 | -0.097 |
| tmem108 | 0.127 | glud1b | -0.095 |
| khdrbs1a | 0.127 | bhmt | -0.094 |
| mnx1 | 0.126 | suclg1 | -0.094 |
| cd9b | 0.126 | sult2st2 | -0.092 |
| CABZ01080702.1 | 0.126 | gstt1a | -0.092 |
| efnb2a | 0.125 | gcshb | -0.091 |
| hmgb3a | 0.125 | tpi1b | -0.091 |
| id1 | 0.125 | lgals2b | -0.090 |
| tpm4a | 0.125 | sod1 | -0.089 |
| ptpro | 0.124 | abat | -0.089 |
| wls | 0.124 | pgk1 | -0.089 |
| mdka | 0.124 | pnp4b | -0.088 |
| hnrnpaba | 0.123 | haao | -0.088 |
| cirbpb | 0.123 | aldh6a1 | -0.088 |
| hmgn6 | 0.123 | agxta | -0.087 |
| aldoaa | 0.123 | rdh1 | -0.087 |
| XLOC-026855 | 0.122 | agxtb | -0.087 |
| tspan7 | 0.121 | upb1 | -0.085 |
| hnrnpabb | 0.121 | tdo2a | -0.085 |
| hmgn7 | 0.121 | grhprb | -0.084 |
| cirbpa | 0.120 | cox7a1 | -0.084 |