family with sequence similarity 169 member Aa
ZFIN 
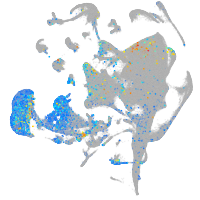
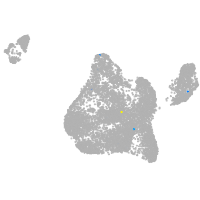
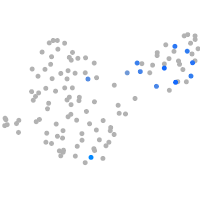
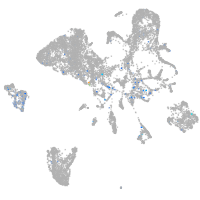
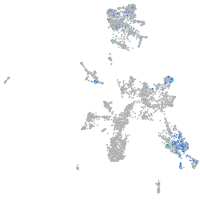
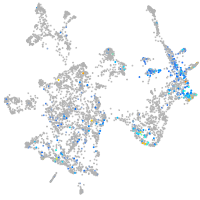
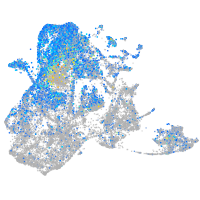
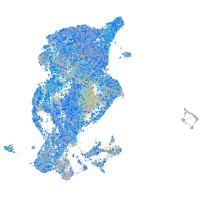
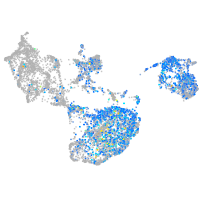
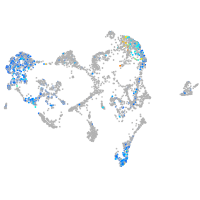
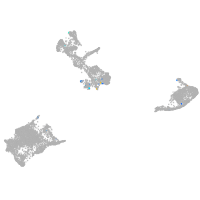
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| abcc8b | 0.305 | gapdh | -0.095 |
| BX004785.3 | 0.238 | gamt | -0.079 |
| rps6ka2 | 0.227 | fbp1b | -0.074 |
| CLIP4 | 0.216 | lgals2b | -0.073 |
| slc1a9 | 0.200 | gatm | -0.072 |
| syt5a | 0.200 | pklr | -0.071 |
| c1qtnf9 | 0.187 | gpx4a | -0.070 |
| si:ch211-157c3.4 | 0.178 | glud1b | -0.065 |
| trim35-21 | 0.177 | pnp4b | -0.063 |
| sv2ba | 0.175 | apoa4b.1 | -0.063 |
| camkvl | 0.170 | apoc2 | -0.062 |
| unc13ba | 0.168 | scp2a | -0.062 |
| CR589947.2 | 0.165 | afp4 | -0.062 |
| krt1-19d | 0.163 | dap | -0.061 |
| CABZ01072096.1 | 0.162 | bhmt | -0.059 |
| ponzr5 | 0.159 | mat1a | -0.059 |
| scel | 0.159 | eno3 | -0.059 |
| dhrs13a.2 | 0.156 | ahcy | -0.059 |
| ANXA1 (1 of many) | 0.155 | apobb.1 | -0.058 |
| asic1a | 0.154 | dhrs9 | -0.058 |
| zgc:193505 | 0.154 | pck1 | -0.056 |
| CR926130.1 | 0.154 | gnmt | -0.055 |
| sh3bp5a | 0.152 | abat | -0.055 |
| mylk4a | 0.146 | cox7a1 | -0.055 |
| anxa1b | 0.145 | tm4sf5 | -0.054 |
| anxa1c | 0.144 | cx32.3 | -0.054 |
| si:ch211-133n4.6 | 0.144 | cx28.9 | -0.054 |
| zgc:194839 | 0.140 | BX908782.3 | -0.053 |
| evpla | 0.140 | gstt1a | -0.053 |
| CABZ01073083.1 | 0.140 | sult2st2 | -0.053 |
| arhgef25b | 0.136 | etnppl | -0.053 |
| si:ch211-76m11.8 | 0.136 | pgam1a | -0.052 |
| CT737131.1 | 0.134 | akr1b1 | -0.052 |
| CABZ01084131.1 | 0.132 | CU682777.2 | -0.052 |
| grp | 0.132 | rbp2a | -0.052 |