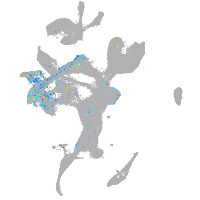
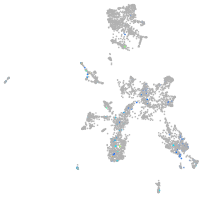
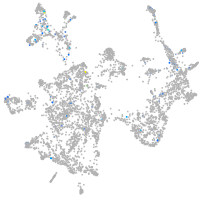
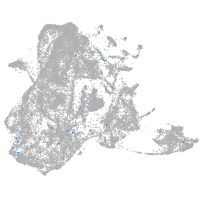
FAM161 centrosomal protein A
ZFIN 
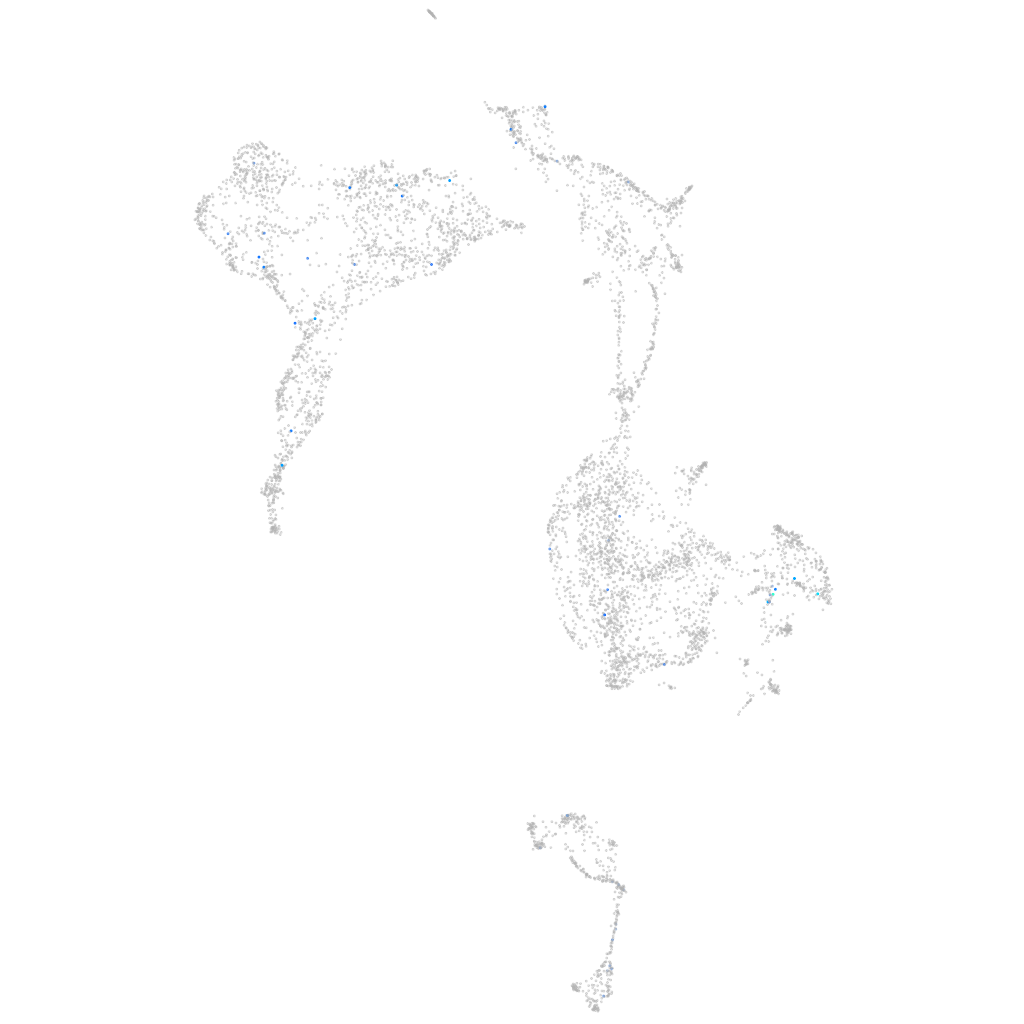
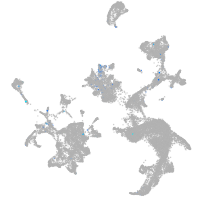
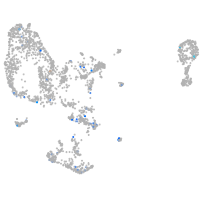
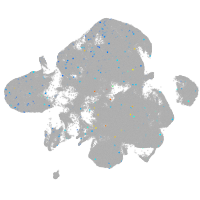
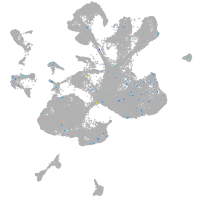
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ankrd33bb | 0.363 | cmn | -0.038 |
| trim35-10 | 0.363 | col2a1b | -0.036 |
| BX323586.3 | 0.257 | plod3 | -0.036 |
| rasgrf2a | 0.255 | loxl1 | -0.035 |
| tm4sf18 | 0.255 | col8a1a | -0.035 |
| kcnj1a.1 | 0.254 | sncga | -0.035 |
| LOC101882645 | 0.250 | colgalt2 | -0.034 |
| zgc:193593 | 0.246 | znf385a | -0.033 |
| XLOC-012427 | 0.246 | sdf4 | -0.032 |
| si:ch73-139e5.4 | 0.231 | hdr | -0.032 |
| XLOC-031884 | 0.223 | cnmd | -0.032 |
| LOC100535225 | 0.221 | gstm.1 | -0.031 |
| hcn4 | 0.221 | ehd2b | -0.031 |
| pimr129 | 0.221 | chst11 | -0.031 |
| si:ch211-160o17.6 | 0.221 | hpcal1 | -0.031 |
| BX088525.2 | 0.219 | calm2b | -0.031 |
| si:ch73-28h20.1 | 0.217 | selenom | -0.030 |
| alkal2b | 0.214 | irf2bp2b | -0.030 |
| glra4b | 0.212 | col5a3a | -0.030 |
| or133-2 | 0.211 | loxl5b | -0.030 |
| LOC103909443 | 0.207 | ssr2 | -0.030 |
| guca1g | 0.199 | tbx3b | -0.029 |
| pdha1b | 0.197 | ca8 | -0.029 |
| tmem237a | 0.194 | susd5 | -0.029 |
| glmnb | 0.194 | fen1 | -0.029 |
| LOC103910570 | 0.194 | fxyd6l | -0.029 |
| CU855758.2 | 0.191 | LOC100534832 | -0.029 |
| dscamb | 0.190 | p3h1 | -0.029 |
| ckmt2a | 0.189 | cavin1b | -0.029 |
| ecscr | 0.185 | matn4 | -0.029 |
| ch25h | 0.184 | hapln1a | -0.029 |
| slc1a2a | 0.183 | col9a3 | -0.029 |
| tmem136b | 0.183 | col4a5 | -0.029 |
| rasip1 | 0.182 | aplp2 | -0.029 |
| zgc:153441 | 0.182 | sema3fb | -0.029 |