fumarylacetoacetate hydrolase (fumarylacetoacetase)
ZFIN 
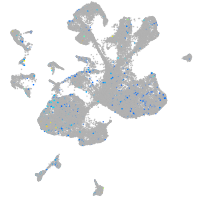
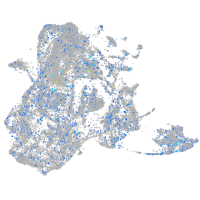
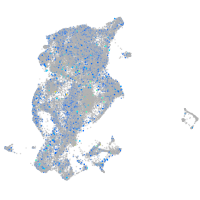
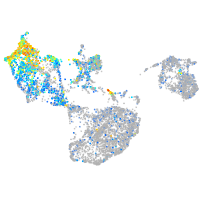
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hpdb | 0.242 | CABZ01075068.1 | -0.047 |
| ttc36 | 0.235 | her15.1 | -0.047 |
| hgd | 0.228 | col11a2 | -0.047 |
| bhmt | 0.185 | otos | -0.043 |
| pcbd1 | 0.183 | epyc | -0.043 |
| gstz1 | 0.178 | tmem88b | -0.042 |
| qdpra | 0.162 | cnmd | -0.042 |
| fmoda | 0.142 | meis3 | -0.042 |
| cbr1l | 0.121 | hbbe3 | -0.041 |
| tat | 0.119 | XLOC-003690 | -0.040 |
| krt18b | 0.117 | hoxb3a | -0.040 |
| gstp1 | 0.115 | col2a1a | -0.039 |
| sod1 | 0.114 | si:ch211-137a8.4 | -0.039 |
| pah | 0.113 | tcf21 | -0.039 |
| msx1b | 0.112 | matn1 | -0.038 |
| aldob | 0.110 | XLOC-003692 | -0.038 |
| rgs5a | 0.109 | her12 | -0.038 |
| si:dkey-16p21.8 | 0.108 | bcam | -0.038 |
| asip2b | 0.107 | gpm6aa | -0.038 |
| krt8 | 0.105 | nr6a1a | -0.038 |
| vwa1 | 0.104 | BX469925.3 | -0.037 |
| twist3 | 0.103 | elavl3 | -0.037 |
| mdh1aa | 0.103 | dla | -0.037 |
| ahcy | 0.103 | marcksl1b | -0.036 |
| mfap2 | 0.100 | rgcc | -0.036 |
| capn12 | 0.099 | nova2 | -0.036 |
| aldh7a1 | 0.098 | wisp3 | -0.036 |
| gstm.1 | 0.098 | lin28a | -0.035 |
| gyg1b | 0.097 | hlx1 | -0.035 |
| dcn | 0.097 | mia | -0.035 |
| fstl3 | 0.096 | tmem108 | -0.035 |
| twist2 | 0.096 | cspg5a | -0.034 |
| ptx3a | 0.095 | rbpms2b | -0.034 |
| aldh6a1 | 0.095 | foxg1a | -0.034 |
| plscr3b | 0.093 | hoxc1a | -0.033 |