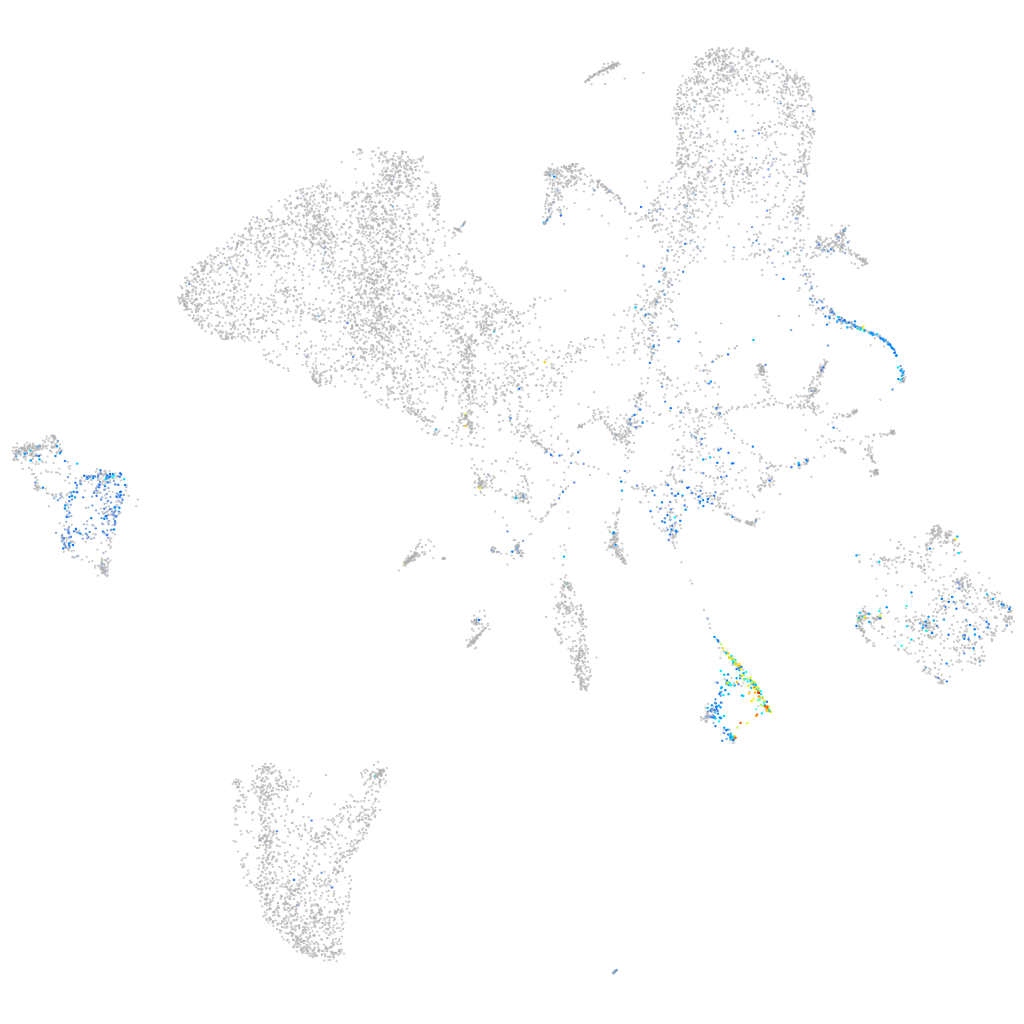
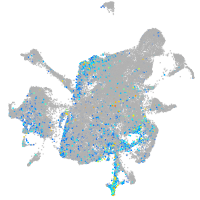
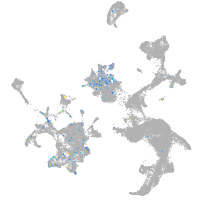
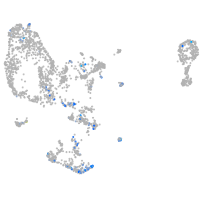
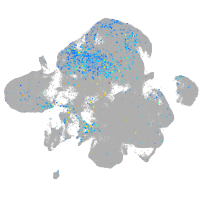
coagulation factor IIIa
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01055365.1 | 0.566 | gapdh | -0.224 |
| pmp22b | 0.561 | gatm | -0.221 |
| ctgfa | 0.551 | gamt | -0.215 |
| sim1b | 0.551 | bhmt | -0.205 |
| mxra8b | 0.528 | fbp1b | -0.197 |
| lama5 | 0.524 | mat1a | -0.195 |
| cabp2b | 0.509 | gpx4a | -0.194 |
| spns2 | 0.508 | apoa4b.1 | -0.192 |
| anxa3b | 0.503 | apoc2 | -0.190 |
| abca12 | 0.502 | ahcy | -0.189 |
| b3gnt5a | 0.493 | afp4 | -0.186 |
| col4a6 | 0.486 | apoa1b | -0.182 |
| cav1 | 0.483 | aqp12 | -0.178 |
| gna15.1 | 0.482 | scp2a | -0.175 |
| bcam | 0.482 | pklr | -0.175 |
| col4a5 | 0.481 | grhprb | -0.174 |
| sftpba | 0.473 | gpd1b | -0.170 |
| sparc | 0.467 | agxtb | -0.170 |
| krt94 | 0.462 | abat | -0.168 |
| pnp4a | 0.462 | gcshb | -0.165 |
| spock3 | 0.458 | mdh1aa | -0.165 |
| ahnak | 0.454 | dap | -0.164 |
| sgms1 | 0.454 | pnp4b | -0.164 |
| vwa1 | 0.454 | apobb.1 | -0.163 |
| spaca4l | 0.453 | hspe1 | -0.162 |
| tgfb3 | 0.450 | aldob | -0.162 |
| amotl2b | 0.446 | kng1 | -0.160 |
| cd151 | 0.445 | apoa2 | -0.159 |
| marcksl1a | 0.440 | suclg1 | -0.159 |
| ceacam1 | 0.439 | agxta | -0.159 |
| col18a1a | 0.432 | si:dkey-16p21.8 | -0.158 |
| tekt3 | 0.430 | rltgr | -0.157 |
| si:ch211-264f5.6 | 0.426 | slc27a2a | -0.157 |
| tmem154 | 0.422 | sult2st2 | -0.156 |
| cavin2b | 0.421 | msrb2 | -0.155 |