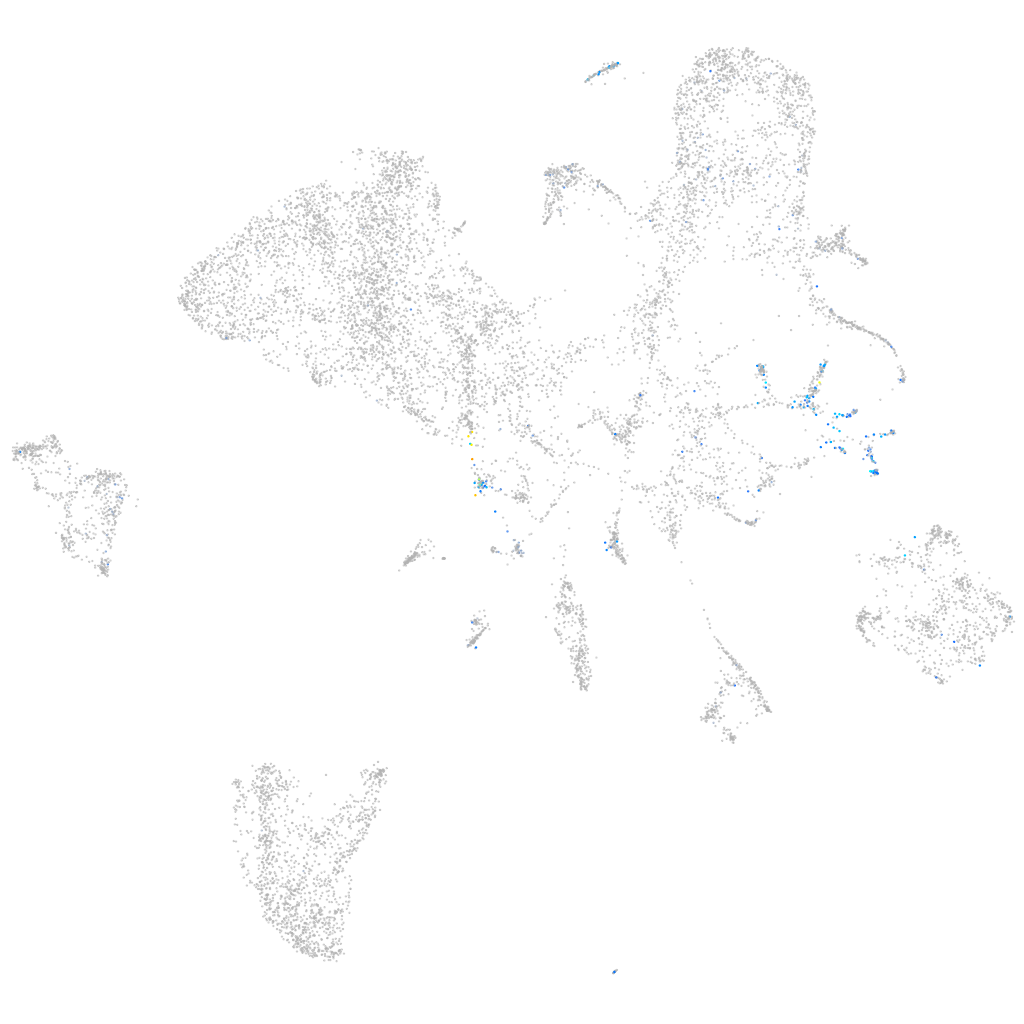
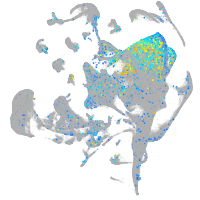
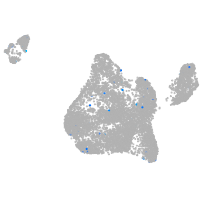
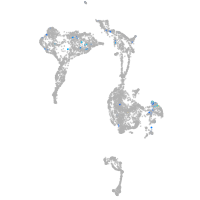
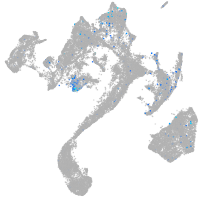
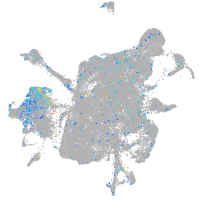
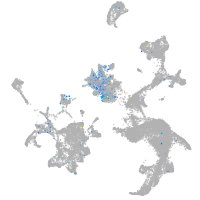
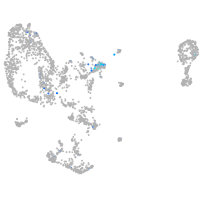
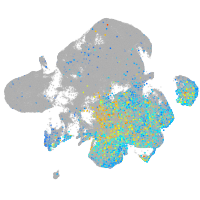
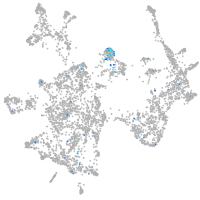
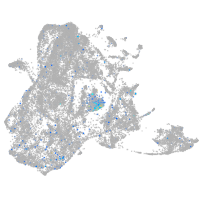
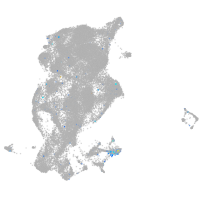
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.321 | ahcy | -0.122 |
| syt1a | 0.296 | gapdh | -0.118 |
| scgn | 0.295 | gamt | -0.111 |
| pcsk1nl | 0.293 | gatm | -0.110 |
| neurod1 | 0.291 | aldob | -0.105 |
| LOC100537384 | 0.285 | nupr1b | -0.099 |
| id4 | 0.285 | fbp1b | -0.098 |
| rnasekb | 0.282 | mat1a | -0.097 |
| evlb | 0.281 | aldh6a1 | -0.093 |
| kcnj11 | 0.281 | bhmt | -0.093 |
| tspan7b | 0.275 | aldh7a1 | -0.093 |
| pax6b | 0.273 | aqp12 | -0.092 |
| atp6v0cb | 0.268 | gstt1a | -0.092 |
| si:dkey-153k10.9 | 0.265 | cx32.3 | -0.091 |
| rasd4 | 0.264 | agxtb | -0.089 |
| cnrip1a | 0.261 | apoa4b.1 | -0.087 |
| mir7a-1 | 0.260 | hdlbpa | -0.086 |
| rprmb | 0.259 | gnmt | -0.086 |
| rims2a | 0.252 | rps20 | -0.085 |
| atpv0e2 | 0.251 | hspe1 | -0.085 |
| gpc1a | 0.247 | rpl4 | -0.085 |
| gapdhs | 0.246 | scp2a | -0.084 |
| cplx2l | 0.246 | pnp4b | -0.084 |
| scg2a | 0.246 | agxta | -0.084 |
| vamp2 | 0.242 | suclg2 | -0.084 |
| insm1a | 0.240 | shmt1 | -0.084 |
| pcsk2 | 0.238 | apoc2 | -0.083 |
| gnao1a | 0.237 | apoa1b | -0.083 |
| vat1 | 0.236 | nipsnap3a | -0.082 |
| kcnk9 | 0.236 | tktb | -0.082 |
| scn3b | 0.234 | si:dkey-16p21.8 | -0.082 |
| tusc3 | 0.234 | aldh1l1 | -0.081 |
| isl1 | 0.234 | ttc36 | -0.081 |
| ndufa4l2a | 0.234 | cebpd | -0.081 |
| eno1a | 0.230 | cdo1 | -0.081 |