ETS variant transcription factor 1
ZFIN 
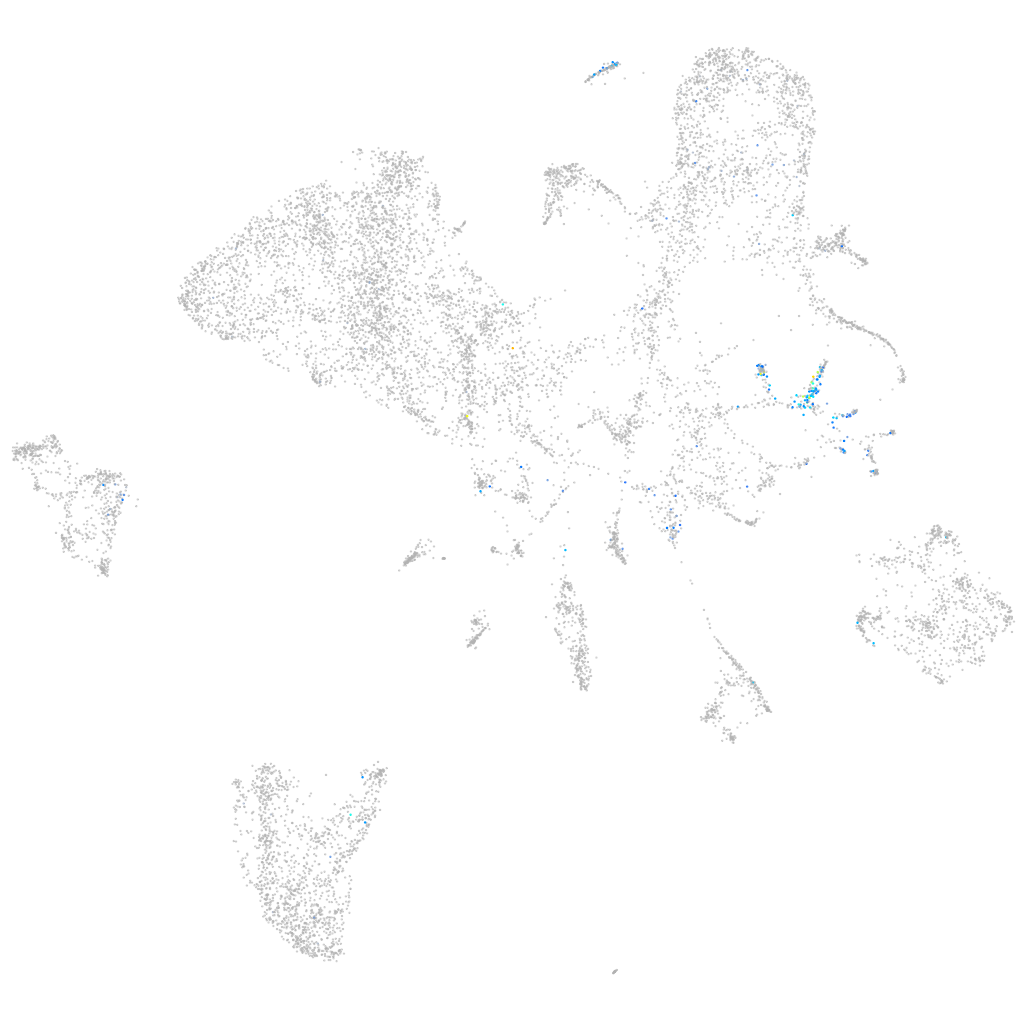
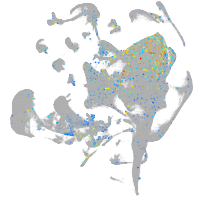
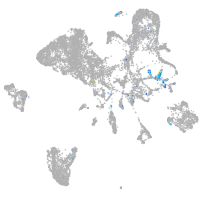
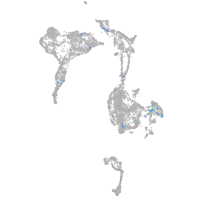
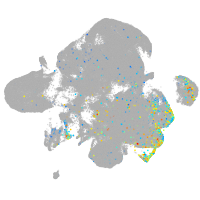
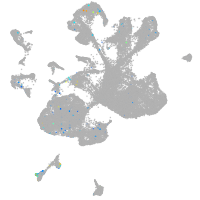
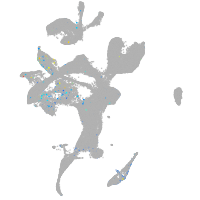
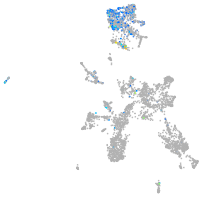
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| arxa | 0.359 | ahcy | -0.109 |
| pyyb | 0.343 | gapdh | -0.101 |
| scg3 | 0.341 | gamt | -0.096 |
| rem2 | 0.332 | gatm | -0.092 |
| rprmb | 0.328 | aldh6a1 | -0.089 |
| rasd4 | 0.328 | fabp3 | -0.087 |
| scgn | 0.328 | si:dkey-16p21.8 | -0.086 |
| neurod1 | 0.322 | bhmt | -0.085 |
| pax6b | 0.306 | hdlbpa | -0.085 |
| ptger3 | 0.305 | mat1a | -0.084 |
| si:ch73-160i9.2 | 0.299 | cx32.3 | -0.083 |
| kcnj11 | 0.295 | aldh7a1 | -0.082 |
| fev | 0.294 | gstr | -0.082 |
| pcsk1nl | 0.291 | rpl6 | -0.081 |
| c2cd4a | 0.290 | fbp1b | -0.080 |
| vat1 | 0.289 | gnmt | -0.080 |
| gfra3 | 0.286 | cebpd | -0.079 |
| egr4 | 0.277 | apoa4b.1 | -0.079 |
| LOC101885494 | 0.260 | agxta | -0.078 |
| dll4 | 0.253 | ppa1b | -0.078 |
| vamp2 | 0.253 | gstp1 | -0.078 |
| capsla | 0.253 | apoa1b | -0.078 |
| hepacam2 | 0.248 | glud1b | -0.077 |
| mir375-2 | 0.248 | hsdl2 | -0.076 |
| mir7a-1 | 0.248 | gstt1a | -0.076 |
| oprd1b | 0.248 | shmt1 | -0.075 |
| si:dkey-153k10.9 | 0.246 | agxtb | -0.074 |
| tspan7b | 0.245 | hspd1 | -0.073 |
| cplx2 | 0.244 | afp4 | -0.073 |
| insm1b | 0.241 | gcshb | -0.073 |
| isl1 | 0.238 | tdo2a | -0.072 |
| nkx2.2a | 0.237 | tktb | -0.072 |
| syt1a | 0.234 | serpinf2b | -0.072 |
| rims2a | 0.233 | wu:fj16a03 | -0.072 |
| camk2n1a | 0.233 | apoc2 | -0.071 |