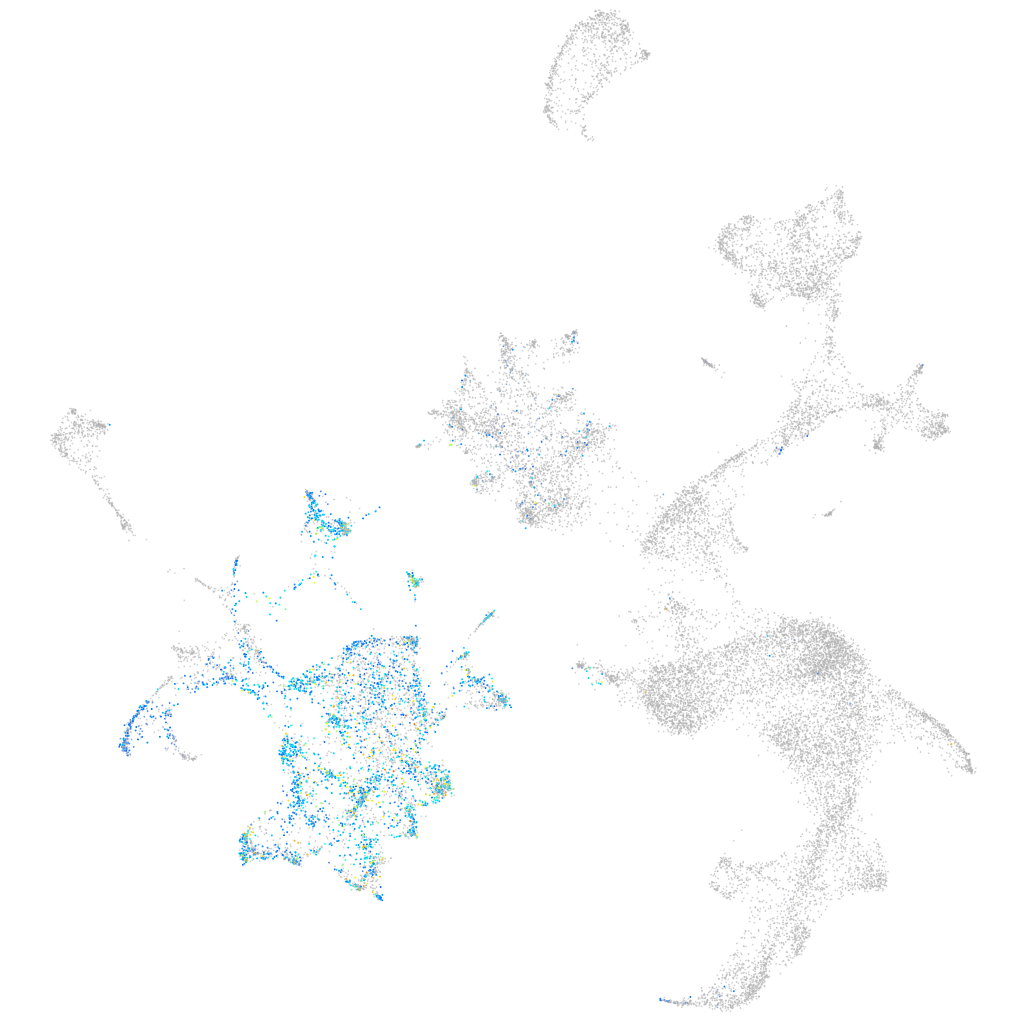
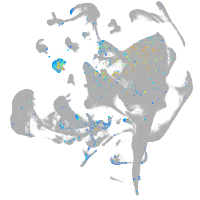
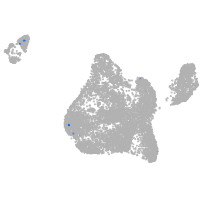
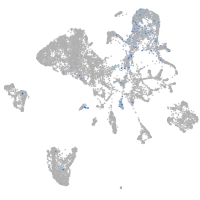
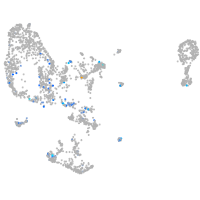
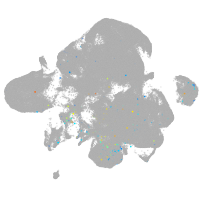
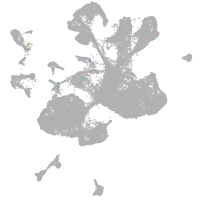
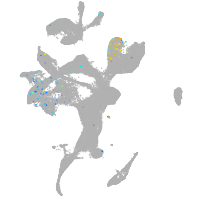
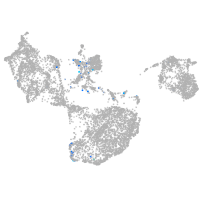
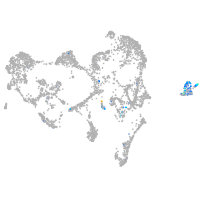
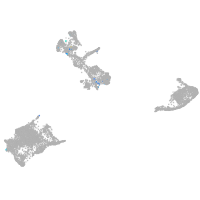
endothelial cell adhesion molecule a
ZFIN 
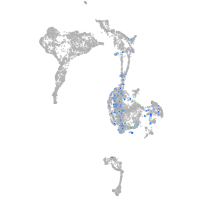
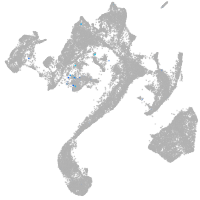
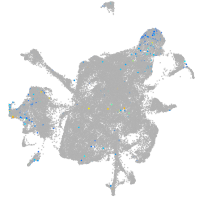
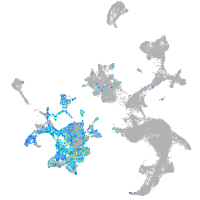
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| krt18a.1 | 0.532 | prdx2 | -0.333 |
| cdh5 | 0.522 | hbae1.1 | -0.321 |
| si:ch211-156j16.1 | 0.516 | hbbe2 | -0.310 |
| clec14a | 0.514 | hbae1.3 | -0.308 |
| tpm4a | 0.513 | hbae3 | -0.306 |
| cldn5b | 0.499 | hbbe1.3 | -0.301 |
| ramp2 | 0.491 | hbbe1.1 | -0.295 |
| egfl7 | 0.491 | cahz | -0.289 |
| si:dkeyp-97a10.2 | 0.487 | blvrb | -0.277 |
| si:dkey-261h17.1 | 0.482 | hemgn | -0.275 |
| krt8 | 0.482 | hbbe1.2 | -0.268 |
| mcamb | 0.470 | fth1a | -0.256 |
| she | 0.458 | alas2 | -0.247 |
| sox18 | 0.457 | slc4a1a | -0.245 |
| myct1a | 0.456 | tspo | -0.244 |
| sox7 | 0.455 | creg1 | -0.243 |
| akap12b | 0.451 | si:ch211-250g4.3 | -0.242 |
| tmem88a | 0.433 | gpx1a | -0.234 |
| dusp5 | 0.432 | nt5c2l1 | -0.234 |
| ecscr | 0.432 | epb41b | -0.220 |
| etv2 | 0.430 | zgc:163057 | -0.218 |
| kdrl | 0.429 | si:ch211-207c6.2 | -0.212 |
| igfbp7 | 0.429 | nmt1b | -0.203 |
| tuba8l3 | 0.424 | plac8l1 | -0.201 |
| msna | 0.424 | tmod4 | -0.198 |
| plk2b | 0.423 | hbae5 | -0.176 |
| tmem108 | 0.421 | tmem14ca | -0.174 |
| cnn3a | 0.419 | klf1 | -0.164 |
| cpn1 | 0.419 | add2 | -0.163 |
| tie1 | 0.416 | si:ch211-227m13.1 | -0.163 |
| crip2 | 0.413 | sptb | -0.160 |
| podxl | 0.412 | si:dkey-240n22.3 | -0.160 |
| cavin2b | 0.408 | tfr1a | -0.159 |
| col4a1 | 0.405 | slc11a2 | -0.156 |
| fam174b | 0.403 | ucp3 | -0.154 |