glutamyl-prolyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-030131-638]
ZFIN 
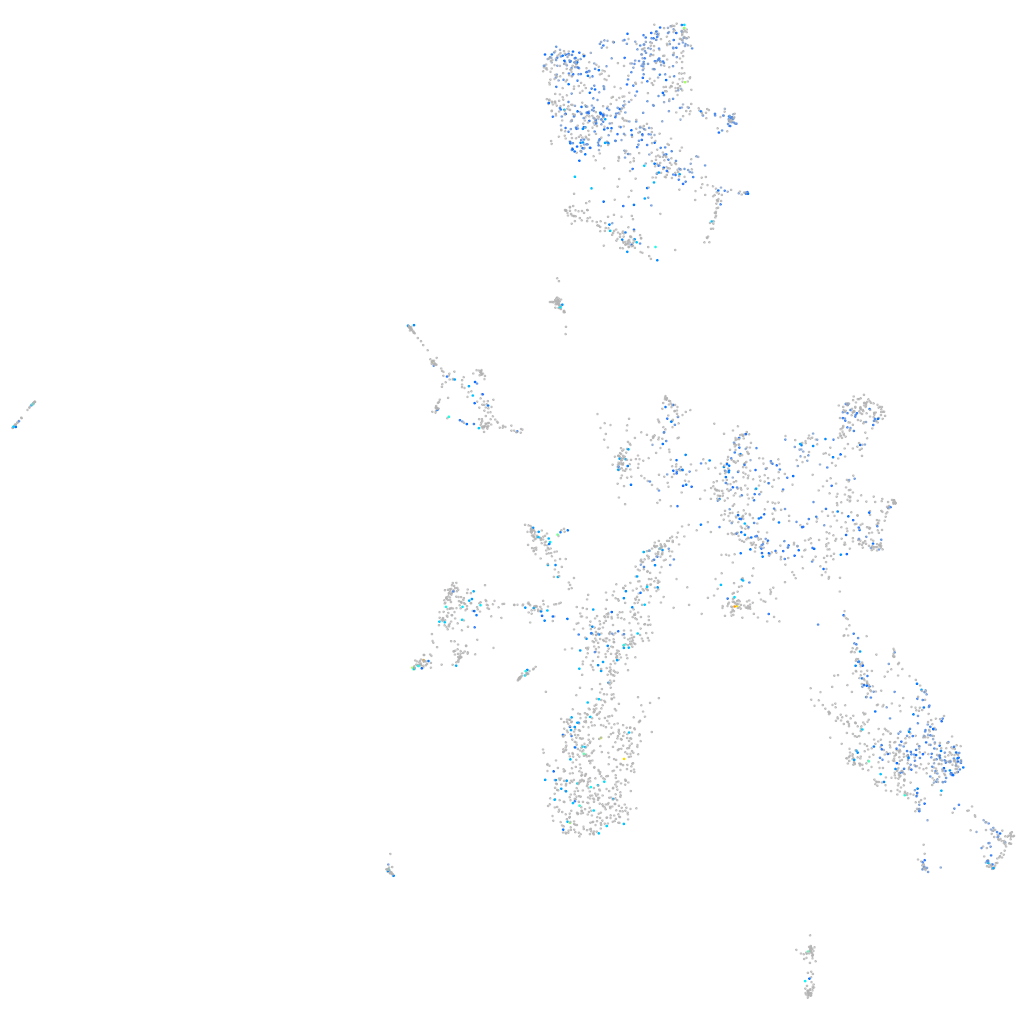
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpr1 | 0.130 | hbbe1.3 | -0.058 |
| hnf1bb | 0.127 | pvalb1 | -0.051 |
| itpkb | 0.116 | desi1a | -0.050 |
| pecam1 | 0.116 | prss59.2 | -0.050 |
| sh3bgrl3 | 0.114 | hbbe2 | -0.049 |
| eif4g1a | 0.114 | tent5ba | -0.047 |
| trpm5 | 0.113 | neurod1 | -0.046 |
| adgrf3b | 0.113 | ppp1r3b | -0.046 |
| si:ch211-218g4.2 | 0.111 | tnnt3a | -0.045 |
| CABZ01072157.1 | 0.111 | hbbe1.2 | -0.045 |
| kcnk17 | 0.111 | CR788248.1 | -0.045 |
| CR318588.3 | 0.110 | elavl3 | -0.044 |
| XLOC-039931 | 0.110 | col2a1b | -0.044 |
| ARF5 (1 of many) | 0.110 | fgf8a | -0.044 |
| si:cabz01007794.1 | 0.109 | LOC101883507 | -0.043 |
| pkd1l3 | 0.109 | ndufa4l2a | -0.042 |
| myo6b | 0.108 | CU467823.1 | -0.042 |
| gpa33a | 0.108 | si:ch211-26b3.4 | -0.042 |
| shha | 0.108 | pvalb2 | -0.041 |
| XLOC-032810 | 0.108 | ptn | -0.041 |
| grtp1a | 0.107 | actc1b | -0.041 |
| tent5bb | 0.106 | adgrl2a | -0.040 |
| perp | 0.105 | ckma | -0.040 |
| ets2 | 0.105 | nme2b.2 | -0.040 |
| igfbp5a | 0.105 | arl15a | -0.040 |
| tubb4b | 0.104 | NC-002333.4 | -0.040 |
| rgs1 | 0.104 | ndrg4 | -0.040 |
| gna14a | 0.103 | tpma | -0.039 |
| abca12 | 0.103 | mapk15 | -0.039 |
| FP016247.2 | 0.103 | LOC108190923 | -0.039 |
| avil | 0.103 | atp1b2b | -0.039 |
| si:ch211-195b11.3 | 0.103 | tspan2a | -0.039 |
| gng13a | 0.103 | dpysl5b | -0.039 |
| ankhd1 | 0.102 | rarres3l | -0.039 |
| mvp | 0.102 | six3b | -0.039 |