epiplakin 1
ZFIN 
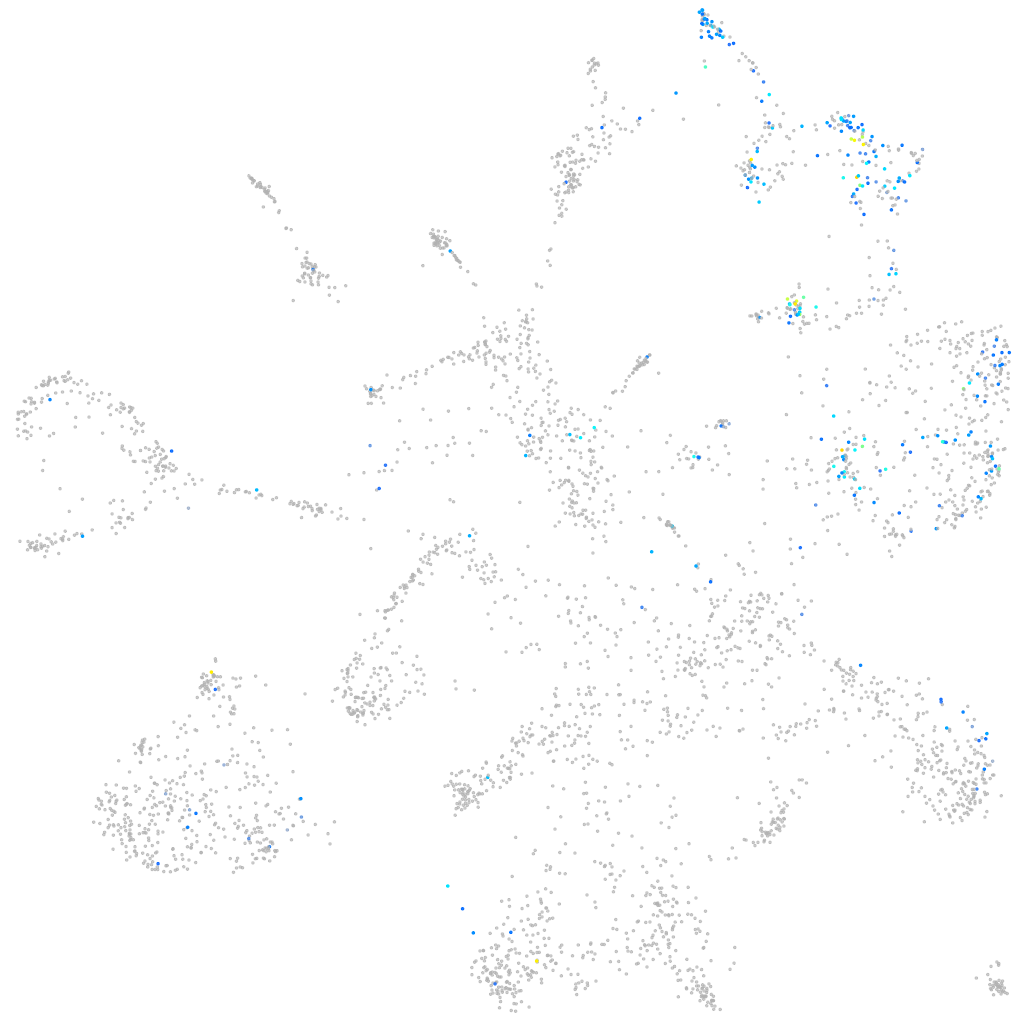
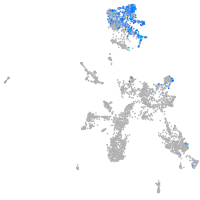
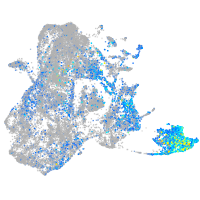
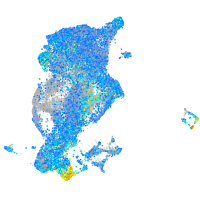
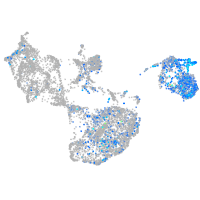
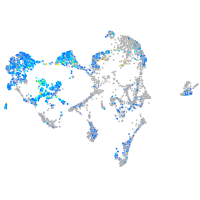
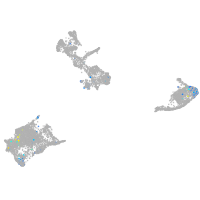
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| podxl | 0.338 | BX088707.3 | -0.193 |
| krt94 | 0.337 | myl9a | -0.170 |
| sfrp5 | 0.312 | tpm1 | -0.151 |
| mmel1 | 0.310 | acta2 | -0.149 |
| synpo2lb | 0.299 | myl6 | -0.143 |
| fabp11a | 0.289 | tagln | -0.140 |
| gstm.3 | 0.285 | myh11a | -0.138 |
| lbx2 | 0.279 | mylkb | -0.130 |
| krt8 | 0.277 | foxf1 | -0.125 |
| tmem88b | 0.276 | lmod1b | -0.120 |
| jam2b | 0.270 | foxf2a | -0.119 |
| bnc2 | 0.264 | ckbb | -0.117 |
| tjp2b | 0.263 | tmem119b | -0.117 |
| gata5 | 0.262 | ptmaa | -0.116 |
| bmper | 0.257 | nkx2.3 | -0.114 |
| cd151 | 0.251 | gapdhs | -0.112 |
| ftr82 | 0.249 | inka1a | -0.111 |
| tgm2b | 0.240 | mdkb | -0.110 |
| wt1b | 0.235 | rbpms2a | -0.109 |
| adma | 0.234 | hmgn2 | -0.107 |
| alpi.1 | 0.232 | cald1b | -0.101 |
| thy1 | 0.230 | csrp1b | -0.100 |
| COLEC10 | 0.229 | ntn5 | -0.100 |
| bcam | 0.226 | fhl2a | -0.097 |
| si:dkey-16l2.20 | 0.225 | ptch2 | -0.096 |
| alx4a | 0.222 | mylka | -0.095 |
| cavin2b | 0.222 | angptl5 | -0.094 |
| jcada | 0.222 | tpm2 | -0.092 |
| lrp2a | 0.219 | BX323087.1 | -0.090 |
| akap12b | 0.219 | gng2 | -0.090 |
| anxa1a | 0.219 | cox6b1 | -0.090 |
| lurap1 | 0.218 | myocd | -0.090 |
| s100a10b | 0.217 | foxp4 | -0.090 |
| tnni1b | 0.216 | XLOC-041870 | -0.089 |
| lmod2a | 0.215 | pcdh18b | -0.089 |