erythropoietin a
ZFIN 
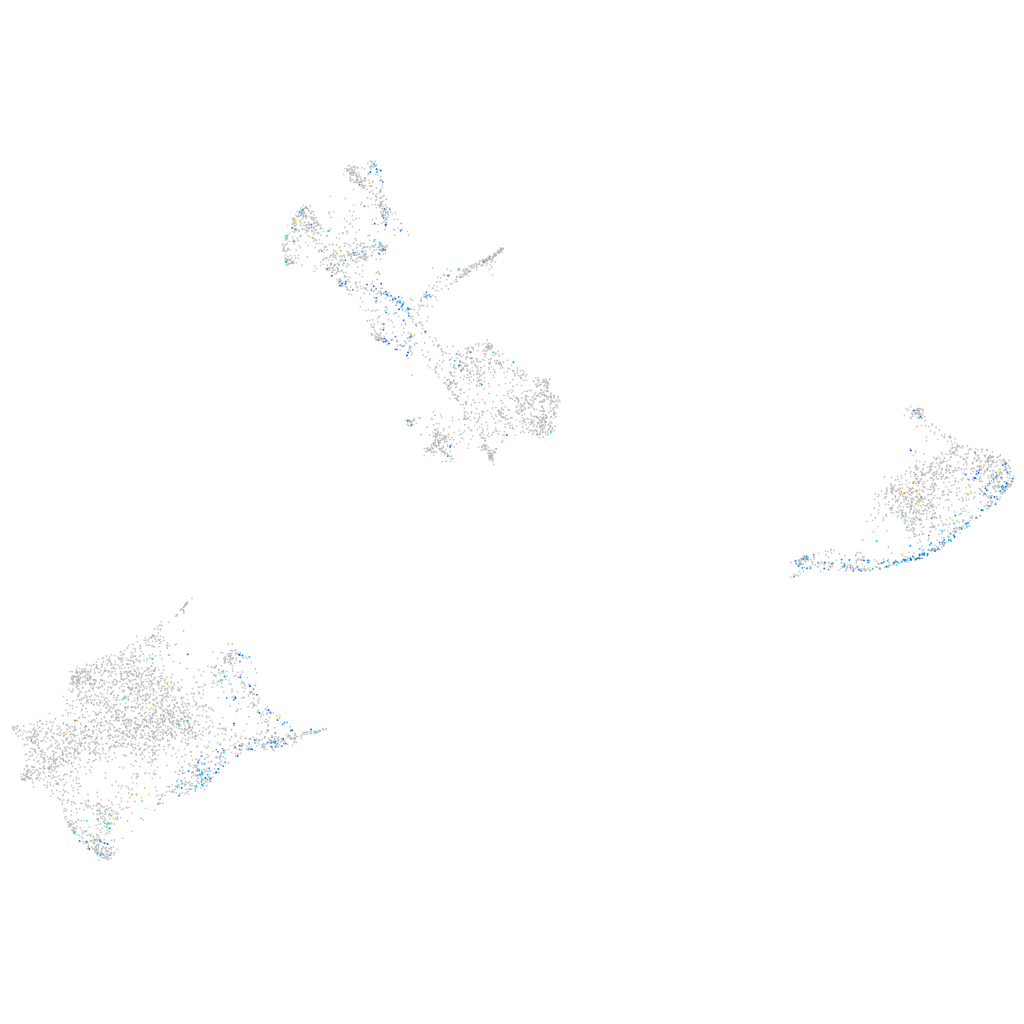
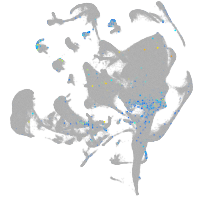
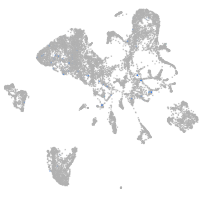
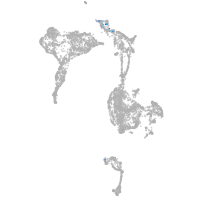
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| msx1b | 0.158 | actc1b | -0.078 |
| naga | 0.151 | pvalb2 | -0.075 |
| mitfa | 0.149 | tuba1c | -0.073 |
| rab32a | 0.145 | CABZ01021592.1 | -0.073 |
| tmem243b | 0.144 | hbae3 | -0.073 |
| si:zfos-943e10.1 | 0.143 | pvalb1 | -0.071 |
| si:ch211-122f10.4 | 0.140 | stmn1b | -0.068 |
| sdc4 | 0.138 | hbbe1.3 | -0.068 |
| tspan36 | 0.137 | ptmaa | -0.065 |
| rabl6b | 0.135 | gpm6aa | -0.063 |
| slc30a7 | 0.130 | hbae1.1 | -0.063 |
| xbp1 | 0.129 | gng3 | -0.063 |
| pim1 | 0.128 | tmsb | -0.061 |
| rgs3a | 0.127 | si:dkey-251i10.2 | -0.061 |
| tram1 | 0.126 | hbbe1.1 | -0.060 |
| slc37a2 | 0.125 | nova2 | -0.059 |
| srsf5a | 0.125 | elavl4 | -0.058 |
| atp6ap1b | 0.125 | uraha | -0.056 |
| atp6v1ba | 0.124 | elavl3 | -0.056 |
| slc3a2a | 0.124 | CU467822.1 | -0.053 |
| gpr143 | 0.123 | mylz3 | -0.051 |
| slc45a2 | 0.123 | krtt1c19e | -0.050 |
| psmb12 | 0.122 | vamp2 | -0.049 |
| degs1 | 0.121 | epb41a | -0.049 |
| qdpra | 0.120 | zgc:110339 | -0.049 |
| tmem9b | 0.120 | tnnt3b | -0.049 |
| pah | 0.119 | sncb | -0.048 |
| syngr1a | 0.119 | myhz1.1 | -0.048 |
| sypl2b | 0.118 | mllt11 | -0.048 |
| pttg1ipb | 0.118 | stx1b | -0.047 |
| scpep1 | 0.117 | sox11a | -0.047 |
| pitpnaa | 0.117 | ndrg2 | -0.047 |
| SPAG9 | 0.117 | hbbe2 | -0.046 |
| osbpl10b | 0.117 | acta1b | -0.046 |
| rab5if | 0.116 | snap25b | -0.046 |