eph receptor A4a
ZFIN 
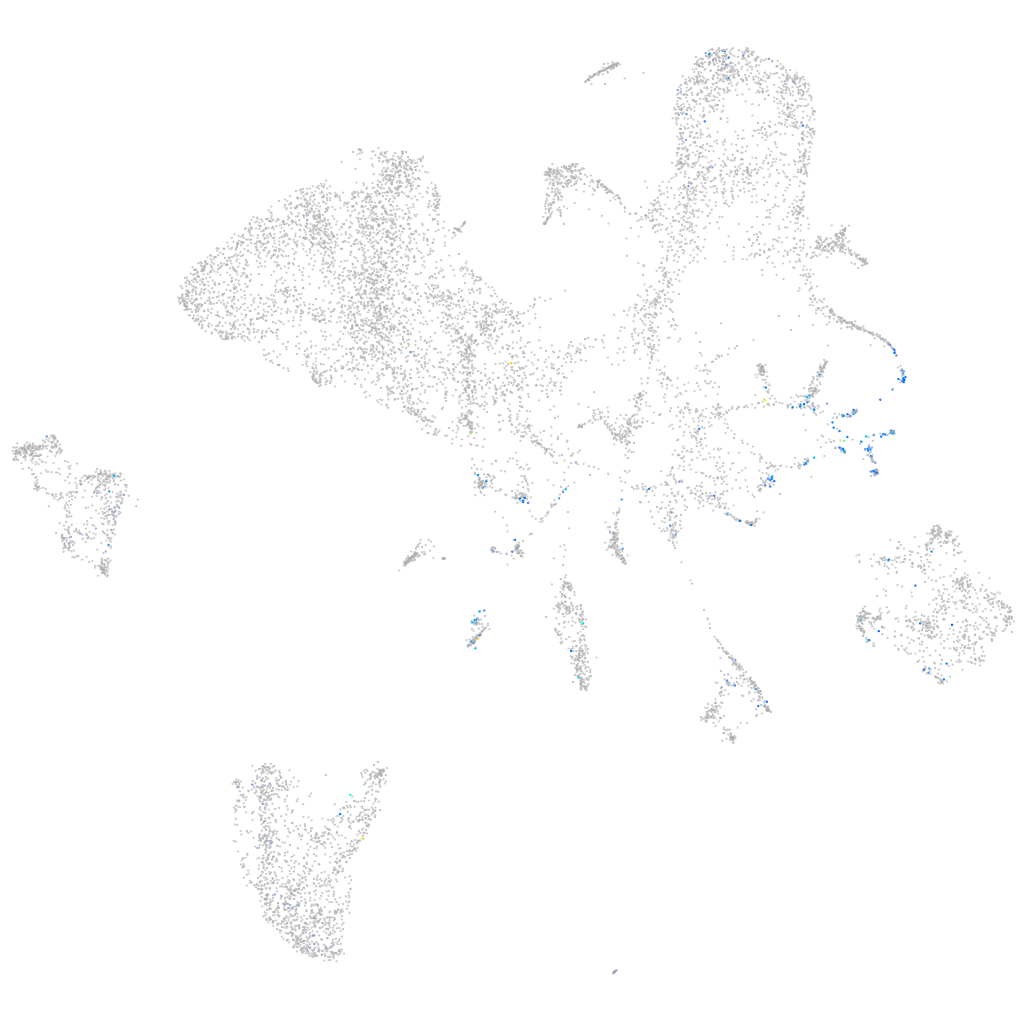
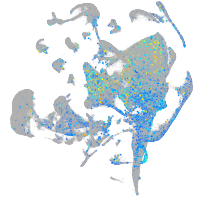
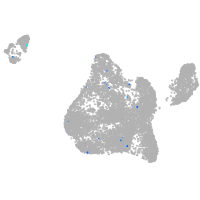
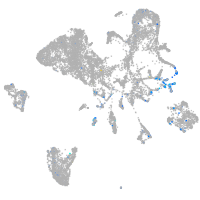
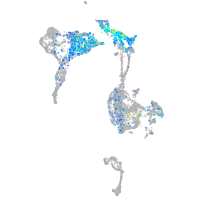
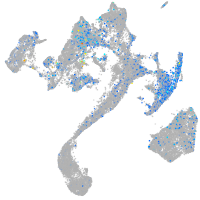
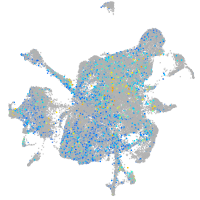
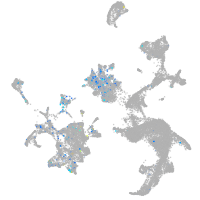
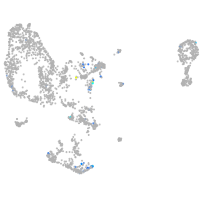
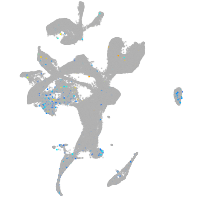
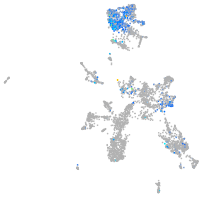
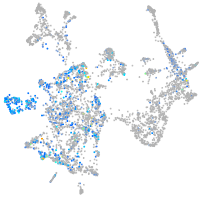
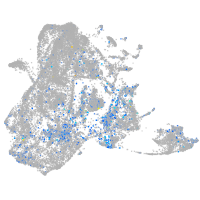
Expression by stage/cluster


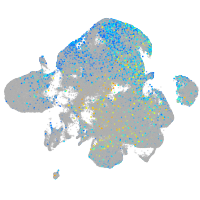
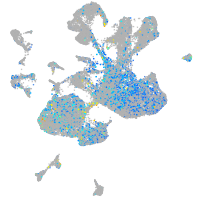
Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kcnj11 | 0.239 | ahcy | -0.120 |
| si:dkey-153k10.9 | 0.232 | aldob | -0.119 |
| pax6b | 0.229 | gapdh | -0.112 |
| scgn | 0.225 | gstt1a | -0.100 |
| isl1 | 0.221 | gamt | -0.100 |
| dkk3b | 0.216 | fbp1b | -0.100 |
| fam49bb | 0.214 | apoa1b | -0.100 |
| rprmb | 0.210 | apoa4b.1 | -0.095 |
| SLC5A10 | 0.205 | mat1a | -0.095 |
| sst2 | 0.197 | tdo2a | -0.094 |
| ndufa4l2a | 0.197 | si:dkey-16p21.8 | -0.093 |
| cadm2a | 0.196 | agxtb | -0.093 |
| scg2a | 0.196 | ftcd | -0.092 |
| CR774179.5 | 0.192 | scp2a | -0.092 |
| gcgb | 0.190 | glud1b | -0.092 |
| scg3 | 0.189 | nipsnap3a | -0.091 |
| mir7a-1 | 0.187 | eno3 | -0.091 |
| elfn1a | 0.185 | apoa2 | -0.089 |
| camk1da | 0.183 | apom | -0.088 |
| slc1a4 | 0.182 | fabp10a | -0.088 |
| pcsk1nl | 0.182 | dpydb | -0.088 |
| sst1.2 | 0.182 | agxta | -0.088 |
| slc35g2b | 0.181 | grhprb | -0.088 |
| neurod1 | 0.179 | haao | -0.087 |
| vat1 | 0.177 | msrb2 | -0.087 |
| renbp | 0.175 | apoc2 | -0.087 |
| kcnk9 | 0.173 | serpina1 | -0.087 |
| tspan7b | 0.173 | gatm | -0.087 |
| syt1a | 0.170 | phyhd1 | -0.086 |
| insm1a | 0.168 | aldh7a1 | -0.086 |
| slc43a2a | 0.168 | sdr16c5b | -0.086 |
| LOC571123 | 0.167 | upb1 | -0.085 |
| gpc1a | 0.167 | si:ch73-361h17.1 | -0.085 |
| foxo1a | 0.165 | fgg | -0.085 |
| tmem145 | 0.165 | gc | -0.085 |