E1A binding protein p300 b
ZFIN 
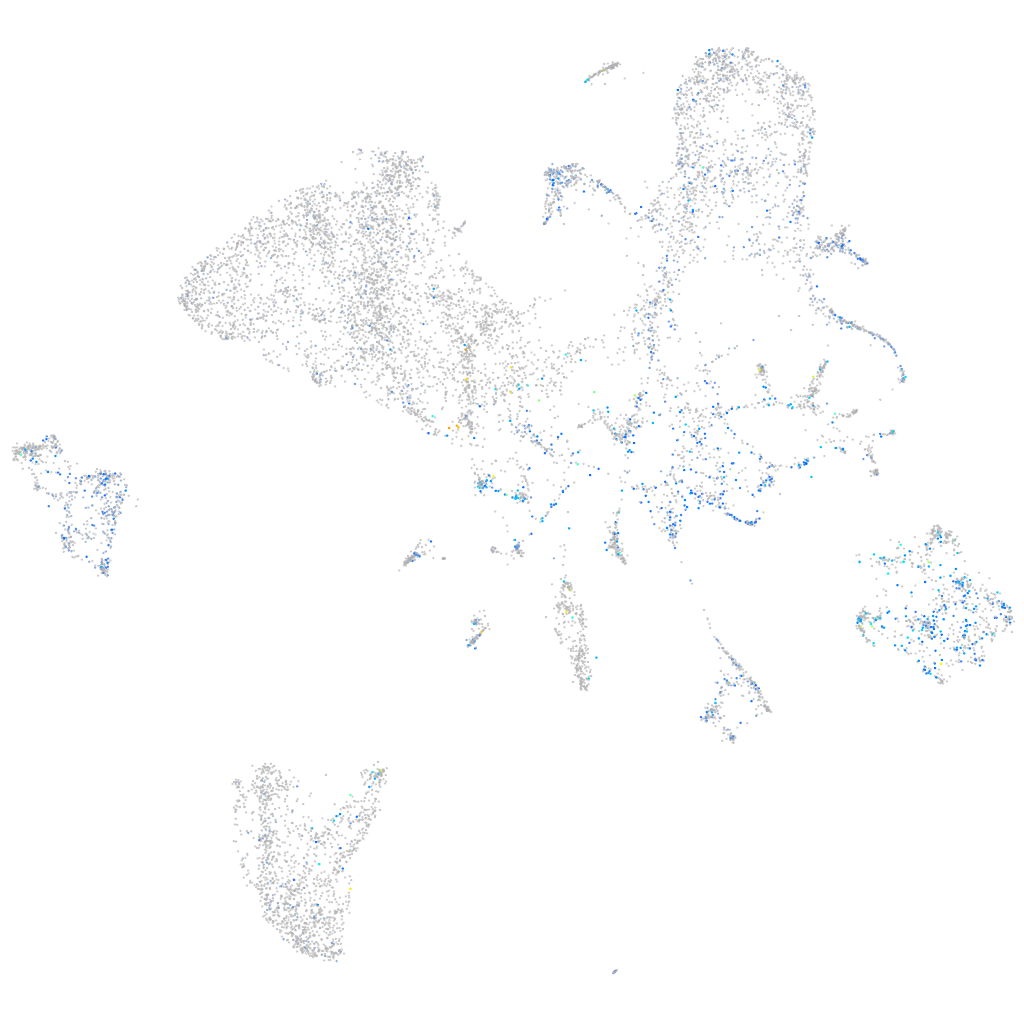
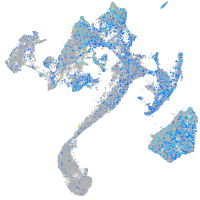
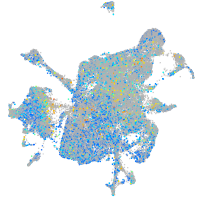
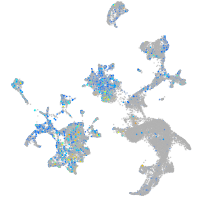
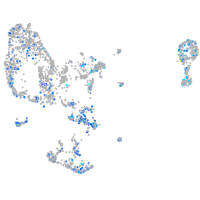
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmgn6 | 0.245 | gamt | -0.225 |
| hmgb3a | 0.240 | gapdh | -0.219 |
| khdrbs1a | 0.234 | ahcy | -0.206 |
| si:ch73-1a9.3 | 0.228 | gatm | -0.200 |
| h3f3d | 0.227 | bhmt | -0.195 |
| si:ch211-222l21.1 | 0.226 | fbp1b | -0.186 |
| marcksb | 0.221 | apoa1b | -0.184 |
| marcksl1b | 0.220 | mat1a | -0.183 |
| hp1bp3 | 0.220 | gpx4a | -0.180 |
| nucks1a | 0.219 | agxtb | -0.178 |
| hnrnpaba | 0.218 | zgc:92744 | -0.176 |
| hnrnpa1a | 0.217 | scp2a | -0.174 |
| cirbpa | 0.216 | glud1b | -0.174 |
| syncrip | 0.216 | apoa4b.1 | -0.173 |
| hdac1 | 0.212 | grhprb | -0.171 |
| acin1a | 0.212 | apoa2 | -0.170 |
| rsf1b.1 | 0.210 | pnp4b | -0.169 |
| ctnnb1 | 0.210 | suclg1 | -0.169 |
| hnrnpa0a | 0.209 | agxta | -0.167 |
| hmga1a | 0.208 | aqp12 | -0.166 |
| ankrd11 | 0.207 | pgm1 | -0.165 |
| si:ch73-281n10.2 | 0.207 | apoc2 | -0.164 |
| ptmab | 0.206 | aldh7a1 | -0.164 |
| h2afvb | 0.206 | eno3 | -0.164 |
| top1l | 0.204 | pklr | -0.163 |
| anp32a | 0.203 | aldob | -0.163 |
| hmgb2a | 0.202 | nupr1b | -0.163 |
| atrx | 0.202 | abat | -0.161 |
| smc1al | 0.202 | ttc36 | -0.161 |
| ubc | 0.201 | kng1 | -0.161 |
| cirbpb | 0.200 | sod1 | -0.161 |
| hnrnpub | 0.199 | serpina1l | -0.161 |
| chd7 | 0.199 | dap | -0.160 |
| cx43.4 | 0.199 | fabp10a | -0.160 |
| hmgb1b | 0.198 | zgc:123103 | -0.160 |