elongin A
ZFIN 
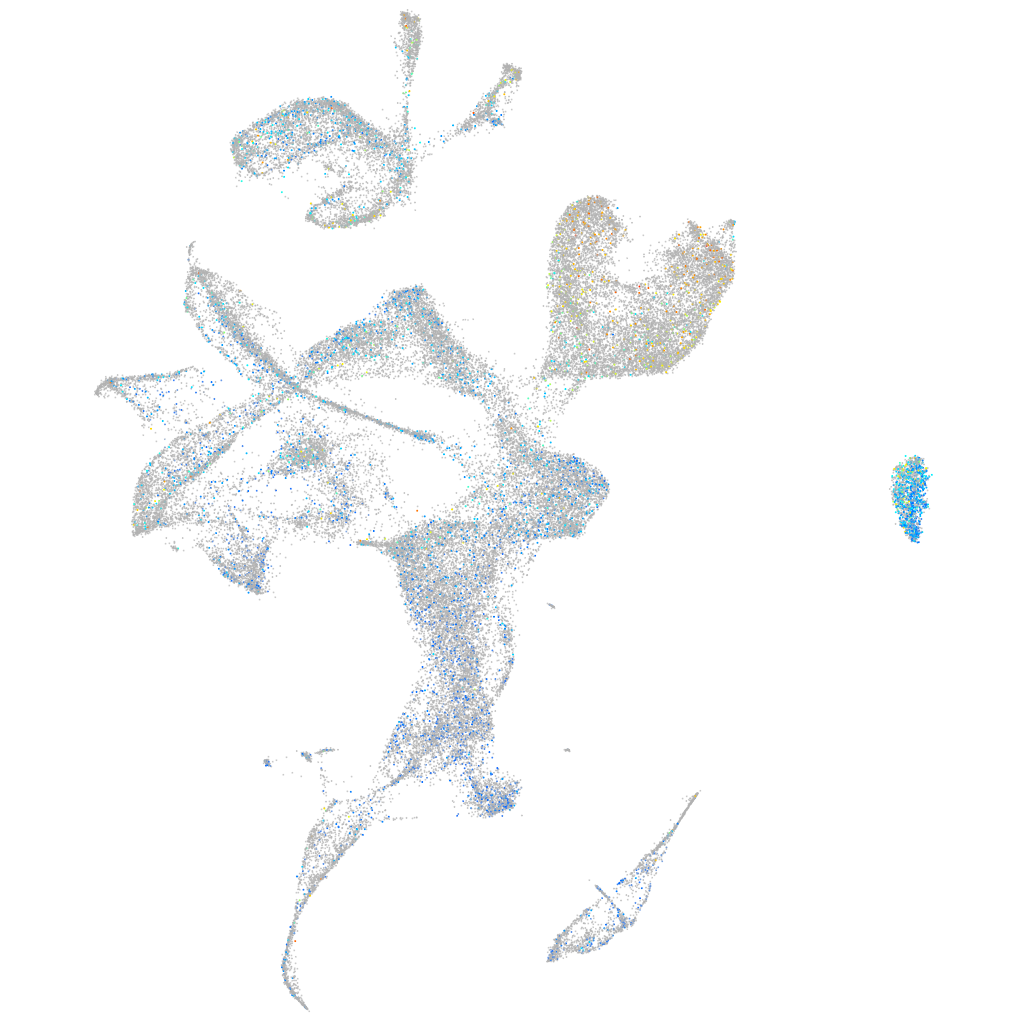
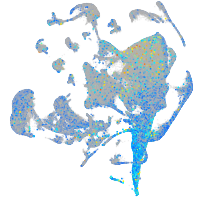
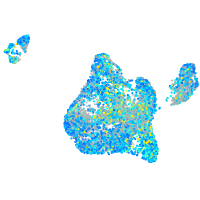
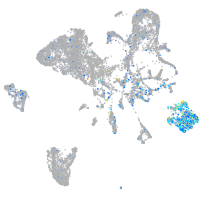
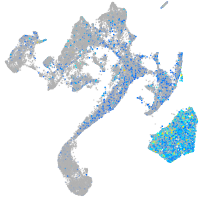
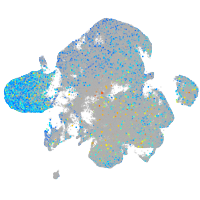
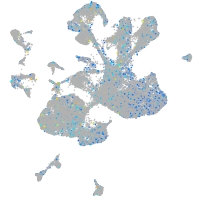
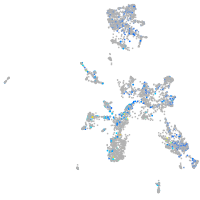
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.166 | rpl37 | -0.111 |
| shisa2a | 0.160 | rps10 | -0.099 |
| stm | 0.157 | zgc:114188 | -0.087 |
| si:ch211-152c2.3 | 0.153 | rps17 | -0.075 |
| hesx1 | 0.145 | h3f3a | -0.071 |
| COX7A2 | 0.145 | zgc:56493 | -0.053 |
| tdgf1 | 0.145 | fabp7a | -0.053 |
| cyp26a1 | 0.144 | ppiab | -0.051 |
| nr6a1a | 0.140 | mt-atp6 | -0.048 |
| apoeb | 0.139 | zgc:158463 | -0.047 |
| sox19a | 0.139 | hnrnpa0l | -0.046 |
| asb11 | 0.135 | ptmaa | -0.046 |
| pkdccb | 0.129 | actc1b | -0.045 |
| LOC108190024 | 0.128 | dut | -0.045 |
| s100a1 | 0.128 | CR383676.1 | -0.043 |
| foxd5 | 0.122 | fabp3 | -0.042 |
| NC-002333.4 | 0.122 | rpl28 | -0.041 |
| polr3gla | 0.119 | COX3 | -0.040 |
| zmp:0000000624 | 0.119 | mt-nd1 | -0.040 |
| apoc1 | 0.118 | nme2b.1 | -0.038 |
| ISCU | 0.117 | prdx2 | -0.038 |
| chrd | 0.115 | mdka | -0.038 |
| alcamb | 0.115 | cct2 | -0.036 |
| aldob | 0.112 | si:ch211-156b7.4 | -0.036 |
| dnmt3bb.2 | 0.107 | rrm2 | -0.036 |
| pprc1 | 0.105 | zgc:165555.3 | -0.035 |
| nnr | 0.105 | COX7A2 (1 of many) | -0.035 |
| si:ch1073-80i24.3 | 0.104 | rps29 | -0.035 |
| pou5f3 | 0.103 | eef1b2 | -0.033 |
| slc2a8 | 0.103 | ahcy | -0.033 |
| bms1 | 0.102 | pvalb2 | -0.033 |
| apela | 0.102 | pvalb1 | -0.033 |
| gnl3 | 0.101 | rpl38 | -0.033 |
| si:dkey-66i24.9 | 0.101 | tuba1c | -0.032 |
| COLGALT1 | 0.100 | hist1h4l | -0.032 |