"eukaryotic translation initiation factor 4, gamma 2a"
ZFIN 
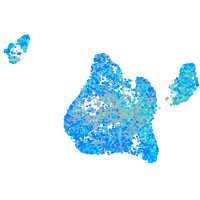
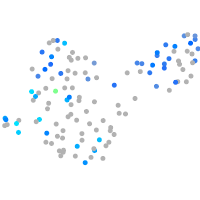
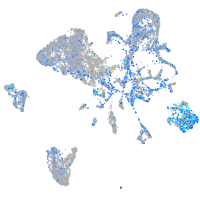
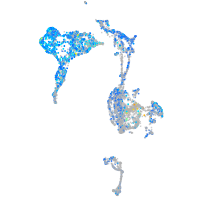
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-56m19.5 | 0.177 | apoa2 | -0.087 |
| hnrnpa1a | 0.165 | dut | -0.084 |
| hnrnpaba | 0.163 | apoa1b | -0.078 |
| hnrnpub | 0.157 | tuba8l | -0.078 |
| hnrnpa0a | 0.157 | prss59.2 | -0.073 |
| sox11b | 0.157 | prss1 | -0.071 |
| chd4a | 0.155 | dap1b | -0.071 |
| myt1a | 0.154 | si:ch211-156b7.4 | -0.070 |
| ddx5 | 0.153 | ctrb1 | -0.065 |
| marcksb | 0.151 | nutf2l | -0.064 |
| elavl3 | 0.150 | stmn1a | -0.064 |
| si:ch211-222l21.1 | 0.149 | gngt2b | -0.063 |
| khdrbs1a | 0.147 | fabp7a | -0.063 |
| cirbpb | 0.145 | pvalb2 | -0.063 |
| tuba1a | 0.145 | rrm2 | -0.062 |
| myt1b | 0.144 | CELA1 (1 of many) | -0.062 |
| pbx4 | 0.144 | pvalb1 | -0.061 |
| ptmab | 0.144 | h1fx | -0.061 |
| tubb5 | 0.143 | lbr | -0.058 |
| ilf2 | 0.142 | krt17 | -0.057 |
| hmgn6 | 0.142 | eef1da | -0.057 |
| smarca4a | 0.142 | prss59.1 | -0.057 |
| dlb | 0.140 | si:dkey-261m9.17 | -0.057 |
| rbm4.3 | 0.139 | selenoh | -0.056 |
| ilf3b | 0.139 | rnaseh2a | -0.056 |
| smarce1 | 0.137 | her2 | -0.056 |
| hdac1 | 0.137 | cks1b | -0.056 |
| tardbp | 0.136 | mdka | -0.055 |
| fscn1a | 0.136 | actc1b | -0.054 |
| nucks1a | 0.133 | zgc:86839 | -0.054 |
| ywhae1 | 0.133 | COX7A2 (1 of many) | -0.053 |
| syncrip | 0.132 | pcna | -0.053 |
| rcc2 | 0.132 | rbp4 | -0.053 |
| hnrnpa0b | 0.130 | myca | -0.053 |
| ebf2 | 0.130 | LOC100330864 | -0.053 |