eukaryotic translation initiation factor 4E nuclear import factor 1
ZFIN 
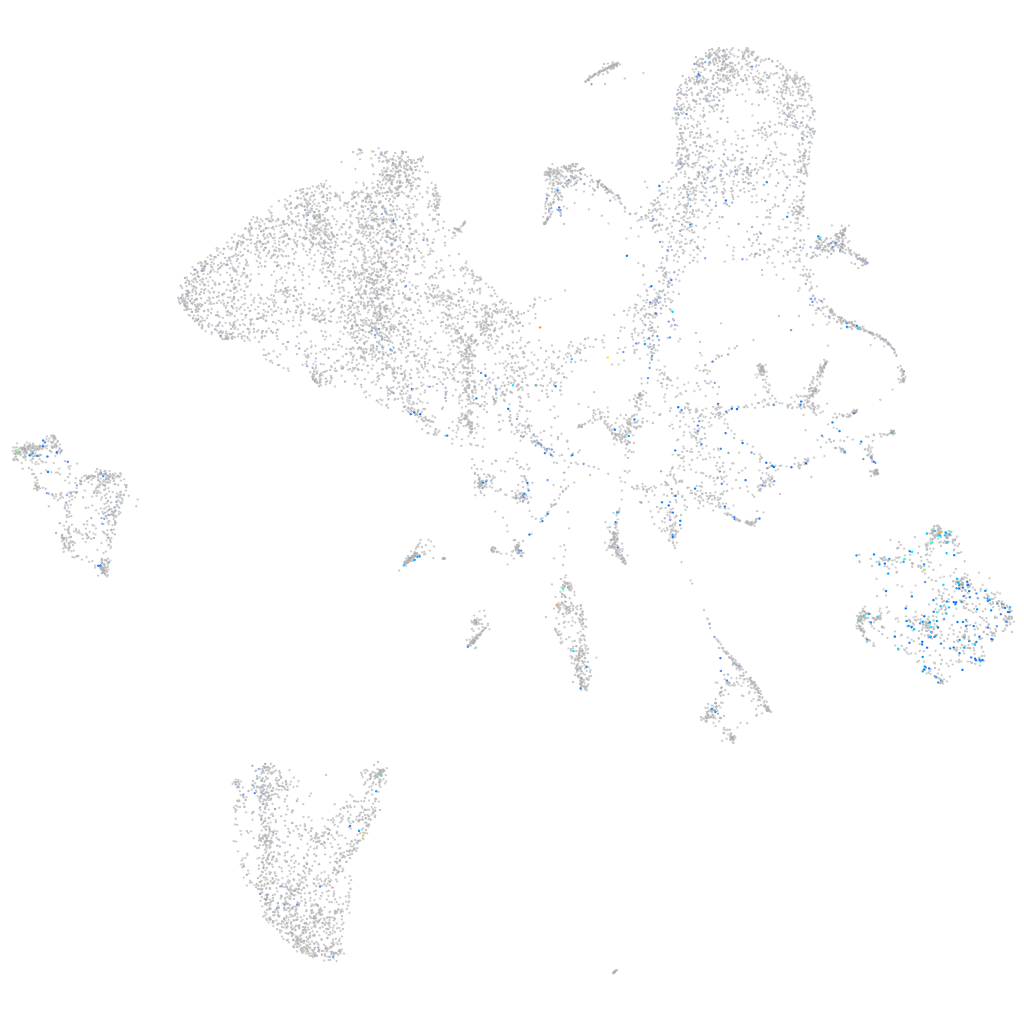
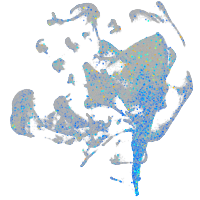
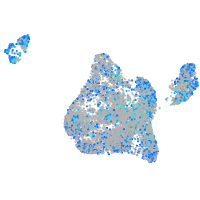
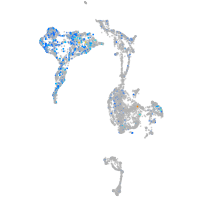
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpa1a | 0.216 | rpl37 | -0.172 |
| hnrnpub | 0.211 | rps10 | -0.171 |
| marcksb | 0.210 | zgc:114188 | -0.148 |
| ppig | 0.208 | gamt | -0.142 |
| hnrnpa1b | 0.206 | rps17 | -0.142 |
| acin1a | 0.206 | ahcy | -0.139 |
| hspb1 | 0.206 | gapdh | -0.130 |
| ilf3b | 0.204 | zgc:92744 | -0.125 |
| nucks1a | 0.203 | nme2b.1 | -0.123 |
| snrnp70 | 0.202 | bhmt | -0.123 |
| brd3a | 0.202 | eno3 | -0.121 |
| seta | 0.201 | apoa1b | -0.120 |
| hmga1a | 0.198 | gatm | -0.119 |
| NC-002333.4 | 0.198 | gpx4a | -0.117 |
| akap12b | 0.197 | suclg1 | -0.117 |
| anp32e | 0.196 | pklr | -0.116 |
| snrpd3l | 0.195 | sod1 | -0.113 |
| si:ch211-152c2.3 | 0.195 | apoa2 | -0.112 |
| hdac1 | 0.194 | fbp1b | -0.112 |
| sf3b2 | 0.191 | apoa4b.1 | -0.111 |
| syncrip | 0.190 | grhprb | -0.110 |
| smo | 0.190 | ckba | -0.110 |
| zgc:110425 | 0.190 | mat1a | -0.109 |
| nop58 | 0.190 | aldob | -0.108 |
| top1l | 0.190 | agxtb | -0.106 |
| hdgfl2 | 0.189 | prdx2 | -0.106 |
| asph | 0.189 | mdh1aa | -0.105 |
| cbx3a | 0.189 | nupr1b | -0.105 |
| marcksl1b | 0.189 | abat | -0.104 |
| srrm2 | 0.188 | afp4 | -0.104 |
| chaf1a | 0.188 | tpi1b | -0.103 |
| hnrnpaba | 0.188 | glud1b | -0.103 |
| khdrbs1a | 0.187 | scp2a | -0.103 |
| AL935174.5 | 0.185 | pnp4b | -0.103 |
| sp5l | 0.185 | rbp4 | -0.102 |