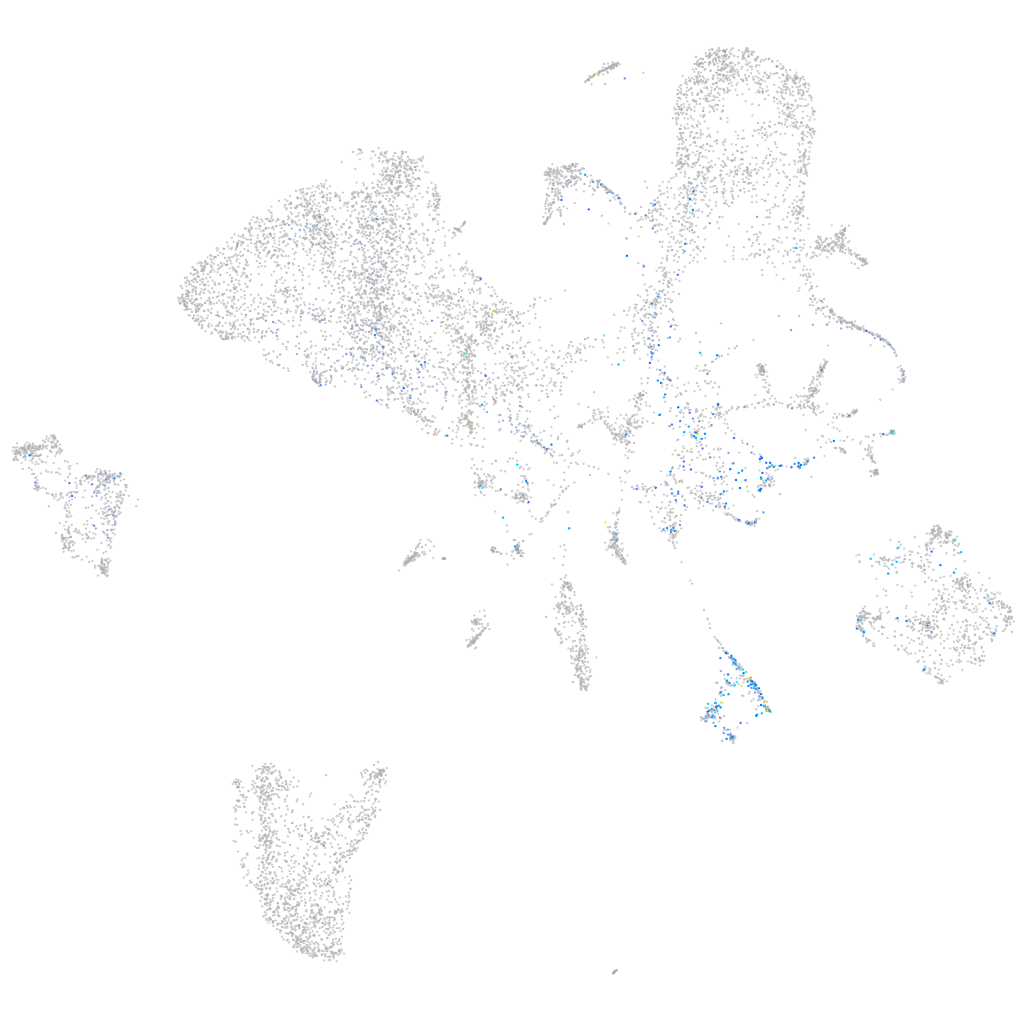
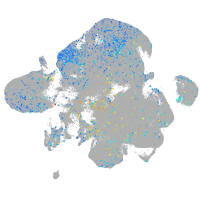
ephrin-B3a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ctgfa | 0.322 | gapdh | -0.132 |
| pmp22b | 0.317 | gatm | -0.124 |
| bcam | 0.312 | ahcy | -0.114 |
| lama5 | 0.312 | gamt | -0.114 |
| sim1b | 0.307 | fbp1b | -0.108 |
| CABZ01055365.1 | 0.290 | dap | -0.101 |
| tekt3 | 0.288 | bhmt | -0.101 |
| cabp2b | 0.285 | aldob | -0.100 |
| spns2 | 0.275 | mat1a | -0.098 |
| marcksl1a | 0.270 | gpx4a | -0.096 |
| abca12 | 0.266 | pnp4b | -0.094 |
| spock3 | 0.265 | apoa4b.1 | -0.090 |
| sftpba | 0.264 | sod1 | -0.088 |
| sim2.1 | 0.263 | afp4 | -0.087 |
| cav1 | 0.262 | apoc2 | -0.086 |
| b3gnt5a | 0.261 | pklr | -0.083 |
| mxra8b | 0.260 | scp2a | -0.083 |
| col4a5 | 0.259 | lgals2b | -0.083 |
| BX663503.3 | 0.255 | mdh1aa | -0.083 |
| amotl2b | 0.254 | abat | -0.083 |
| arhgap29b | 0.253 | pck1 | -0.082 |
| gna15.1 | 0.253 | cox7a1 | -0.081 |
| sim2 | 0.252 | akr1b1 | -0.080 |
| FP102018.1 | 0.252 | apobb.1 | -0.080 |
| anxa3b | 0.250 | agxtb | -0.080 |
| f3a | 0.250 | apoa1b | -0.079 |
| col4a6 | 0.245 | slc16a10 | -0.078 |
| col18a1a | 0.241 | rdh1 | -0.078 |
| sparc | 0.240 | suclg2 | -0.078 |
| cygb1 | 0.239 | grhprb | -0.078 |
| sgms1 | 0.238 | gnmt | -0.077 |
| lsp1 | 0.237 | ucp1 | -0.077 |
| agrn | 0.237 | g6pca.2 | -0.077 |
| pnp4a | 0.235 | sult2st2 | -0.076 |
| si:ch211-264f5.6 | 0.231 | eno3 | -0.076 |