eukaryotic translation elongation factor 1 gamma
ZFIN 
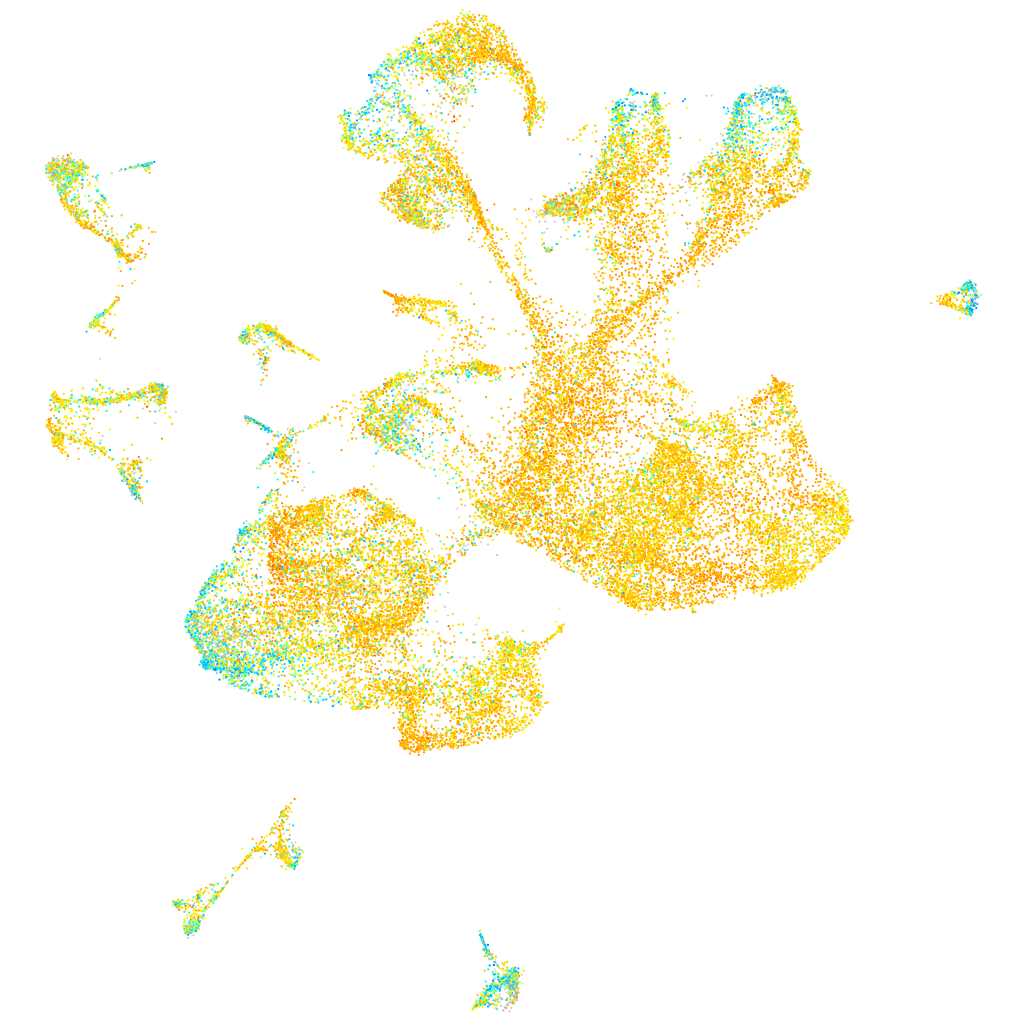
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rps5 | 0.513 | glula | -0.310 |
| rpl19 | 0.512 | slc1a2b | -0.297 |
| rps20 | 0.503 | cx43 | -0.293 |
| rpl10 | 0.502 | slc4a4a | -0.289 |
| rpl8 | 0.501 | acbd7 | -0.287 |
| rps7 | 0.501 | aldocb | -0.279 |
| rpl7a | 0.501 | atp1a1b | -0.273 |
| rpl13 | 0.498 | mt2 | -0.265 |
| rps3a | 0.498 | gapdhs | -0.259 |
| rps9 | 0.497 | slc6a1b | -0.257 |
| rplp0 | 0.496 | ptn | -0.254 |
| rpl21 | 0.494 | efhd1 | -0.251 |
| rps27.1 | 0.494 | gpr37l1b | -0.244 |
| rps23 | 0.494 | ppap2d | -0.229 |
| rpl17 | 0.492 | slc38a3a | -0.227 |
| rpl10a | 0.488 | ndrg3a | -0.227 |
| rps4x | 0.487 | pygmb | -0.224 |
| rpl7 | 0.486 | mfge8a | -0.219 |
| eef1b2 | 0.484 | cyp2ad3 | -0.218 |
| rpl13a | 0.483 | slc3a2a | -0.217 |
| rack1 | 0.482 | ptgdsb.2 | -0.217 |
| hsp90ab1 | 0.482 | ckbb | -0.213 |
| rpsa | 0.481 | cdo1 | -0.211 |
| rps14 | 0.481 | cahz | -0.210 |
| rpl11 | 0.481 | IGLON5 | -0.206 |
| rps2 | 0.478 | smox | -0.205 |
| rps27a | 0.477 | mgll | -0.203 |
| rps12 | 0.477 | fabp7a | -0.200 |
| faua | 0.476 | FO704813.1 | -0.200 |
| naca | 0.476 | ak1 | -0.199 |
| rps19 | 0.473 | cspg5b | -0.198 |
| rps15a | 0.472 | CR383676.1 | -0.195 |
| rpl18 | 0.472 | si:ch211-66e2.5 | -0.195 |
| eef1a1l1 | 0.471 | cox4i2 | -0.194 |
| rpl12 | 0.470 | eno1a | -0.192 |