endothelin receptor type Ab
ZFIN 
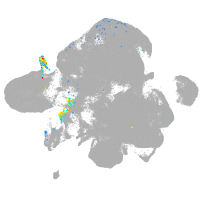
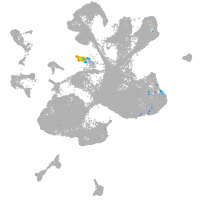
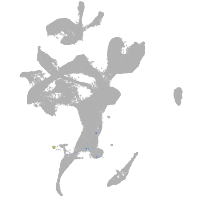
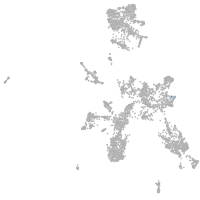
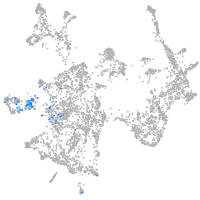
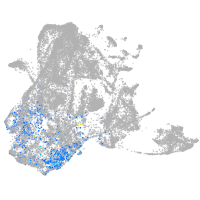
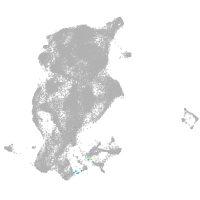
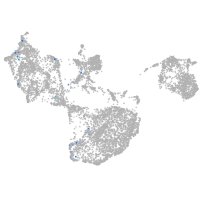
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcdh2ab5 | 0.337 | aldob | -0.055 |
| FP016239.1 | 0.318 | dap | -0.047 |
| trpm2 | 0.305 | sod1 | -0.046 |
| tnnc1a | 0.294 | gapdh | -0.044 |
| XLOC-022924 | 0.293 | ahcy | -0.042 |
| LOC108183529 | 0.290 | fbp1b | -0.042 |
| LO018260.1 | 0.286 | BX908782.3 | -0.041 |
| LOC110438280 | 0.278 | atp5f1d | -0.040 |
| XLOC-010093 | 0.273 | eno3 | -0.040 |
| CR847844.2 | 0.268 | gstt1a | -0.040 |
| ccdc80l1 | 0.257 | hadh | -0.039 |
| nppa | 0.255 | pklr | -0.039 |
| LOC103910193 | 0.241 | pgam1a | -0.038 |
| LOC110439013 | 0.240 | lgals2b | -0.038 |
| XLOC-036926 | 0.236 | slc25a20 | -0.038 |
| LOC110437924 | 0.232 | atp5mc3b | -0.038 |
| gpib | 0.204 | gstp1 | -0.037 |
| pgm5 | 0.198 | rmdn1 | -0.037 |
| cmlc1 | 0.198 | mt-atp6 | -0.037 |
| LOC110438542 | 0.196 | etfa | -0.037 |
| BX470183.1 | 0.192 | mat1a | -0.037 |
| c1s | 0.191 | scp2a | -0.037 |
| smtnl1 | 0.189 | sod2 | -0.037 |
| obsl1b | 0.183 | cox6a1 | -0.036 |
| BX511250.1 | 0.180 | suclg2 | -0.036 |
| LOC110439489 | 0.179 | ugt1a7 | -0.036 |
| CU041319.1 | 0.178 | glud1b | -0.036 |
| CABZ01062996.1 | 0.175 | ddt | -0.035 |
| lrrc3cb | 0.173 | suclg1 | -0.035 |
| gcm2 | 0.172 | nupr1b | -0.035 |
| mitfa | 0.170 | sdr16c5b | -0.035 |
| BX901974.1 | 0.169 | gnmt | -0.035 |
| prkg1b | 0.169 | atp5if1b | -0.035 |
| BX682558.1 | 0.168 | aco1 | -0.035 |
| tfpi2 | 0.162 | abat | -0.035 |