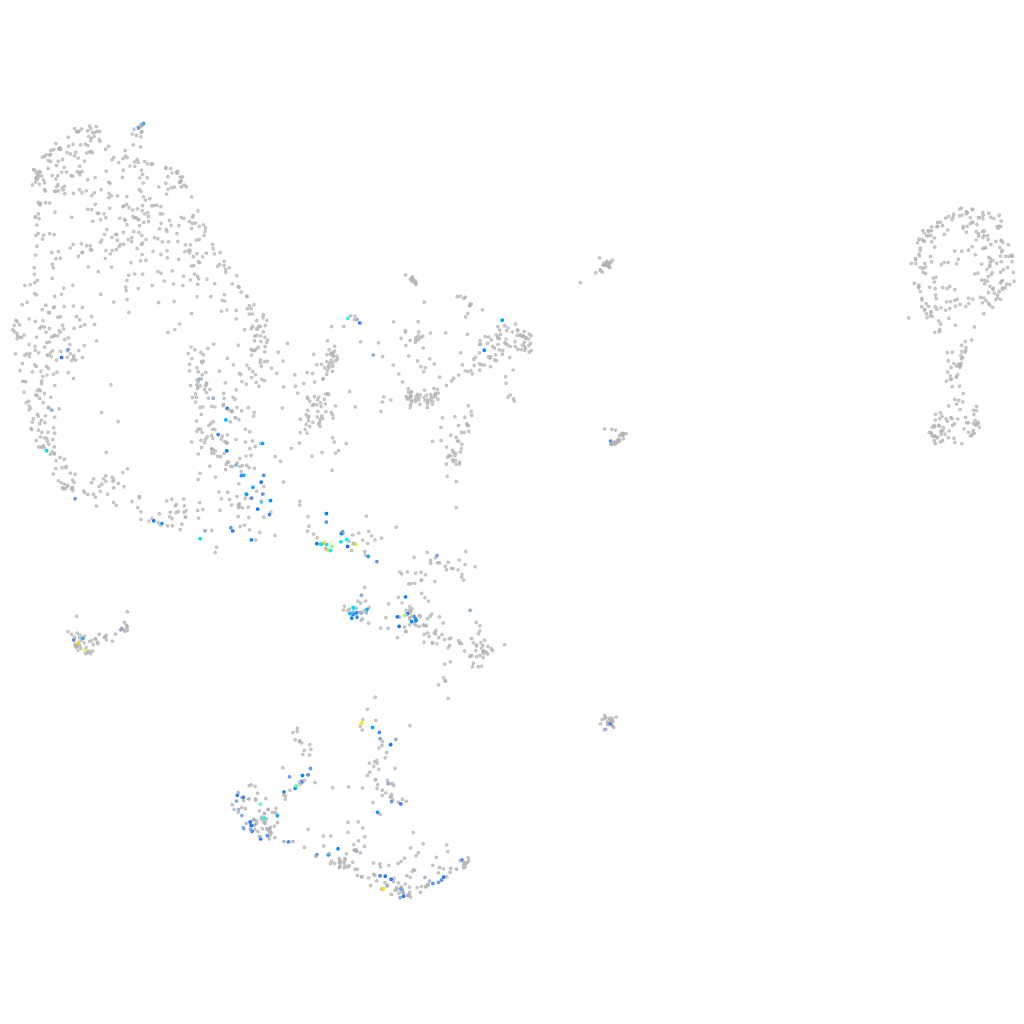
endothelin 2
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 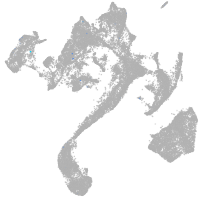 |
mural cells / non-skeletal muscle  |
mesenchyme 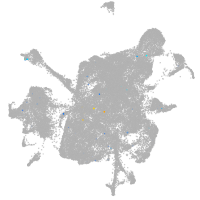 |
hematopoietic / vasculature 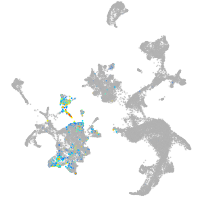 |
pronephros 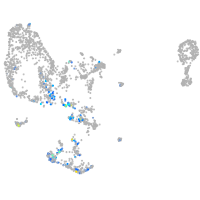 |
neural |
spinal cord / glia |
eye |
taste / olfactory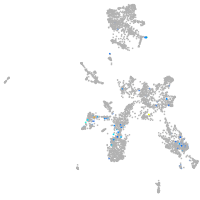 |
otic / lateral line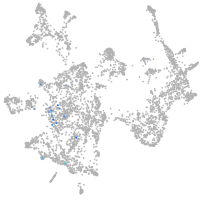 |
epidermis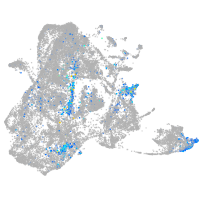 |
periderm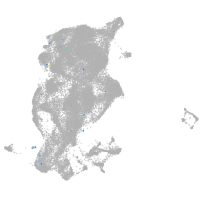 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:64022 | 0.329 | dpys | -0.135 |
| elf3 | 0.284 | alpl | -0.134 |
| wnt2ba | 0.275 | hao1 | -0.132 |
| si:dkey-23i12.7 | 0.270 | si:dkey-33i11.9 | -0.131 |
| gadd45bb | 0.266 | gamt | -0.126 |
| enkur | 0.263 | gpt2l | -0.124 |
| atf3 | 0.263 | pnp6 | -0.124 |
| cldnb | 0.259 | gapdh | -0.120 |
| daw1 | 0.251 | aco1 | -0.119 |
| ppdpfa | 0.251 | nit2 | -0.119 |
| pim1 | 0.246 | hoga1 | -0.118 |
| nme5 | 0.243 | grhprb | -0.118 |
| zgc:158343 | 0.243 | epdl2 | -0.118 |
| si:ch211-226m7.4 | 0.241 | haao | -0.118 |
| ctgfa | 0.239 | nipsnap3a | -0.117 |
| nipa2 | 0.238 | fbp1b | -0.116 |
| phlda3 | 0.236 | cx32.3 | -0.116 |
| krt18a.1 | 0.235 | gstt1a | -0.116 |
| mycbp | 0.234 | g6pca.2 | -0.115 |
| krt8 | 0.233 | agxtb | -0.114 |
| efcab1 | 0.232 | glyctk | -0.113 |
| gmnc | 0.228 | cubn | -0.111 |
| si:dkeyp-75h12.7 | 0.227 | pdzk1ip1 | -0.111 |
| iqcg | 0.226 | phyhd1 | -0.110 |
| morn3 | 0.225 | adh8b | -0.109 |
| cd9b | 0.225 | ctsz | -0.108 |
| myl9b | 0.224 | gcshb | -0.107 |
| cdh6 | 0.222 | rgn | -0.107 |
| ccdc173 | 0.221 | etnppl | -0.106 |
| id2b | 0.217 | ahcy | -0.105 |
| cldnc | 0.217 | pklr | -0.104 |
| efhc1 | 0.217 | slc22a13b | -0.104 |
| mir21-1 | 0.215 | upb1 | -0.104 |
| ribc2 | 0.215 | dhrs9 | -0.103 |
| her6 | 0.214 | si:ch211-201h21.5 | -0.103 |